Summary
Fanconi anemia (FA) is an autosomal recessive disorder exhibiting chromosomal fragility, bone-marrow failure, congenital abnormalities, and cancer. At least eight complementation groups have been described, with group A accounting for 60%–65% of FA patients. Mutation screening of the group A gene (FANCA) is complicated by its highly interrupted genomic structure and heterogeneous mutation spectrum. Recent reports of several large deletions of FANCA, coupled with modest mutation-detection rates, led us to investigate whether many deletions might occur in the heterozygous state and thus fail to be detected by current screening protocols. We used a two-step screening strategy, in which small mutations were detected by fluorescent chemical cleavage of the FANCA transcript, and heterozygosity for gross deletions was detected by quantitative fluorescent multiplex PCR. We screened 26 cell lines from FA complementation group A for FANCA mutations and detected 33 different mutations, 23 of which were novel. Mutations were observed in all 26 cell lines and included 43 of a possible 52 mutant alleles (83%). Of the mutant alleles, 40% were large intragenic deletions that removed up to 31 exons from the gene, indicating that this may be the most prevalent form of mutation in FANCA. Several common deletion breakpoints were observed, and there was a highly significant correlation between the number of breakpoints detected in a given intron and the number of Alu repeats that it contained, which suggests that Alu-mediated recombination may explain the high prevalence of deletions in FANCA. The dual screening strategy that we describe may be useful for mutation screening in other genetic disorders in which mutation-detection rates are unexpectedly low.
Introduction
Fanconi anemia (FA; MIM 227650) is an autosomal recessive disorder associated with a wide range of clinical features, including progressive bone-marrow failure, diverse developmental abnormalities, and a high incidence of acute myeloid leukemia (Auerbach et al. 1993). FA cells are hypersensitive to DNA cross-linking agents, such as diepoxybutane (DEB) and mitomycin C (MMC), and this provides a valuable laboratory test to support the clinical diagnosis. The disorder is genetically heterogeneous, with at least eight complementation groups described to date (Joenje et al. 1997). This indicates that FA may be caused by a defect in at least eight different genes, which have been denoted “FANCA”–“FANCH.” The FANCA, FANCC, and FANCG genes have been cloned (Strathdee et al. 1992; Fanconi Anemia Breast Cancer Consortium 1996; Lo Ten Foe et al. 1996; de Winter et al. 1998), and the FANCD and FANCE genes have been mapped to chromosomes 3p and 6p, respectively (Whitney et al. 1995; Waisfisz et al. 1999). FANCA is the most commonly mutated FA gene, accounting for 60%–65% of all cases.
Mutation screening of the FANCA gene is a difficult task, since the 4.3-kb coding sequence is interrupted by 43 exons (Ianzano et al. 1997), and the mutational spectrum is very heterogeneous. More than 100 different FANCA mutations have been described to date and include almost all classes of mutations (Fanconi Anemia Breast Cancer consortium 1996; Lo Ten Foe et al. 1996; Levran et al. 1997; Savino et al. 1997; Tachibana et al. 1999; Wijker et al. 1999). Mutation-detection rates in these studies are ∼30%–70%, and there have been several recent reports of intragenic deletions in FANCA (Centra et al. 1998; Levran et al. 1998; Wijker et al. 1999). This led us to postulate that the low mutation-detection rates might be caused by a high frequency of intragenic deletions, which would not be detected by the majority of screening protocols if they occurred in compound heterozygotes whose other mutant allele was a point mutation. We therefore developed a two-step fluorescence-based mutation-screening strategy that would detect both small mutations in the coding sequence of FANCA and heterozygous deletions of one or more exons. The method was then used to screen the FANCA gene for mutations in 26 cell lines from patients with FA who were classified as group A by complementation analysis. Mutations were observed in all 26 cell lines and included a high frequency of intragenic deletions, some of which removed >30 exons from the FANCA gene. This strategy may have general application for mutation screening in other disorders.
Patients and Methods
Patients
Patients were ascertained by EUFAR (European Concerted Action on Fanconi Anemia Research), and blood samples were obtained with informed consent. Twenty-six unrelated FA patients had been diagnosed previously, on the basis of their clinical presentation and their cellular hypersensitivity to mitomycin C (MMC) or diepoxybutane (DEB). Lymphoblastoid cell lines were established and tested by complementation analysis to identify group A patients, as described elsewhere (Joenje et al. 1995).
Methods
RNA Extraction and Reverse-Transcriptase PCR (RT-PCR)
Total RNA was extracted from lymphoblasts, as described elsewhere (Chomczynski and Sacchi 1987). From each cell line, 2.5 μg of total RNA was reverse transcribed by use of 25 ng each of the following specific primers, in order to ensure transcription of the entire coding sequence: 1727R (5′-GCCTCCATGACGGTGACTGG-3′), 3′ R1 (5′-GCTCCAGGTCAGCTACCATCTCCTGC-3′), 3494R (5′-GAAGTCGACCATCAGGGAGGG-3′), and RU4955R (5′-CAGAGCTGGACTTTGCCTGT-3′).
These primers were multiplexed in a single reaction and heated for 10 min at 65°C. Then, to each patient sample, 1 × reverse transcriptase buffer, 10 mM DTT, 500 μM each dNTP, 40 U RNase inhibitor, and 200 U MMLV reverse transcriptase (Gibco BRL) was added, incubated at 42°C for 1 h, and heated at 95°C for 3 min to destroy the enzyme. The full coding sequence was amplified in six overlapping fragments, with ⩾200 bp of overlapping sequence between each pair of fragments. A primary PCR was carried out for each of the six fragments, with 0.4 μM each of forward and reverse primer (for primer sequences and conditions, see table 1), 500 μM each dNTP, 1 × Taq DNA polymerase buffer (67 mM Tris-HCl, pH 8.8, 16.6 mM (NH4)2SO4, X mM MgCl2, where X is 1.5–6.7, 170 μg BSA/ml, 10 mM β-mercaptoethanol), 1 μl cDNA, and 1.5 U (Promega) Taq polymerase. PCR conditions were an initial denaturation step at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 10 s, annealing for 30 s, and extension for 1 min at 72°C for each kilobase of fragment size. A final extension for 5 min at 72°C was added. Nested PCR was then carried out by addition of 1 μl of the primary PCR product to a secondary PCR mix, as before (see table 1), but with the inclusion of 0.3 μl of one of three different fluorescently-labeled dUTP solutions: TAMRA-yellow, R6G-green, or R110-blue (PE Biosystems). Secondary products were then electrophoresed in a 1.5% agarose gel to check for aberrantly sized bands, which were Geneclean-purified and sequenced by use of the ABI 377 DNA sequencer. Full-size fragments were used directly for fluorescent chemical cleavage of mismatch (FCCM) analysis (see below). Probes used for FCCM were amplified from the FANCA cDNA (Lo Ten Foe et al. 1996), as for patient cDNA, but with use of biotinylated primers as well as fluorescent dUTPs.
Table 1.
Primer Pairs for Six Overlapping RT-PCR Fragments of FANCA Gene
|
PCR Conditions |
||||||
| Fragment | 1o and 2o PCR Primer Pairs (5′→3′) | Nucleotide Positiona | Exons Spanned | Product Size (bp) | Annealing Temperature (°C) | [Mg2+] of PCR (mM) |
| 1 | Forward, CCA AGG CCA TGT CCG ACT CG | 13–595 | 1–6 | 581 | 60 | 1.5 |
| Reverse, CAG AAA GCA TGG CCC TGG CGA CG | ||||||
| Forward, GGG TCC CGA ACT CCG CCT CGG GCC | 55 | 1.5 | ||||
| Reverse, TTT CCA GCA GCT CTT GCA GG | ||||||
| 2 | Forward, GCG CCT CCT GCG AAG CCA TCA G | 393–1079 | 4–12 | 686 | 66 | 1.5 |
| Reverse, CAG CTG GCA GCT CTC GAA TGC | ||||||
| Forward, CAG TCA CCC TGT GCT GCT GAC | 64 | 1.5 | ||||
| Reverse, CGG TAC AGT GAG GTG AGC AG | ||||||
| 3 | Forward, CAG ATC TGA GAA GAA CTG TGG | 893–1627 | 11–18 | 734 | 60 | 1.5 |
| Reverse, GCC TCC ATG ACG GTG ACT GG | ||||||
| Forward, GGT TCG GAG TGT TCA GTG GAC | 60 | 1.5 | ||||
| Reverse, GCT CAG TAA TGT CCC CAG CTG | ||||||
| 4 | Forward, GGT TCG GAG TGT TCA GTG GAC | 1100–1900 | 13–21 | 800 | 60 | 5.0 |
| Reverse, CAT CAC GCT GGC TGG GGT CTG | ||||||
| Forward, GTG CAG AGG AGT TGG TTG G | 60 | 4.0 | ||||
| Reverse, CTT CTG GCT TCT CTT CAG CAG | ||||||
| 5 | Forward, CAT CAT GGT GTT TGA GCA TAC G | 1724–3308 | 19–33 | 1,584 | 60 | 6.7 |
| Reverse, GAA GTC GAC CAT CAG GGA GGG | ||||||
| Forward, GGA GGC CTT ACT ACG TGT CC | 60 | 6.7 | ||||
| Reverse, GCA GTG ATG GGC TGT TCT GCC TG | ||||||
| 6 | Forward, GCA AGA CCT CAT AGT GCC TC | 3093–4486 | 32–3′ UTR | 1,327 | 60 | 6.7 |
| Reverse, CGC AAA CGC TGA GTG ACT CG | ||||||
| Forward, CAG ACG GCT CCA GGC TCT GAC | 60 | 6.7 | ||||
| Reverse, CCC ACT AAA GCA GTC GAG G | ||||||
Numbering, from the first nucleotide of the initiation codon.
Solid-Phase FCCM Detection
This procedure was based on the use of chemical cleavage to detect mismatched bases in heteroduplexes formed between wild-type and patient DNA (Cotton et al. 1988; Ellis et al. 1998). The original method has been modified to a solid-phase, fluorescence-based protocol that allows rapid and sensitive detection of point mutations and other small lesions (Rowley et al. 1995). Two microliters of biotinylated, fluorescently labeled probe was gel-purified and added to 2 μl fluorescently labeled target in 1 × hybridization buffer (0.3 M NaCl, 100 mM Tris-HCl, pH 8.0) in a total volume of 30 μl. This was denatured for 5 min at 95°C, then held for 60 min at 65°C to form heteroduplexes, which could be stored at 4°C for several hours. Streptavidin-coated beads (Dynal) were treated by taking 10 μl of beads for every patient sample DNA and washing twice in 1 × binding and washing buffer (5 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 M NaCl), before resuspension in 20 μl 2 × binding and washing buffer. This 20-μl bead suspension was then added to the hybrid DNA and resuspended. After 15 min the supernatant was removed from the beads by use of a magnet (Promega). The beads with the bound DNA were then resuspended in 20 μl hydroxylamine solution (4 M hydroxylamine hydrochloride, 2.3 M diethylamine) and incubated at 37°C for 2 h or were resuspended in osmium tetroxide solution (0.4% OsO4, 2% pyridine) and incubated at room temperature for 15 min. The supernatant was removed from the beads as before, and the beads were washed in binding and washing buffer before resuspension in 5 μl piperidine solution (2 M piperidine in formamide plus dextran blue dye) and heating at 90°C for 15 min. Two microliters of the supernatant was loaded on a 6% acrylamide denaturing gel and electrophoresed for 5 h on an ABI 377 DNA sequencer. Data were collected and analyzed by means of Genescan 2.0 ABI 377 software (see electrophoretogram, in fig. 1).
Figure 1.
Electrophoretogram showing FCCM analysis of fragment 3 in patient BD237. Both this sample and a control show alternative splicing of exon 14. Rox2500=MW marker.
Quantitative Fluorescent Multiplex PCR
Genomic DNA was prepared from lymphoblastoid cell lines by salt/chloroform extraction, as described elsewhere (Gibson et al. 1996). DNA samples were then incubated at 55°C for 1 h; half of this sample was taken to determine the concentration, and the remainder was diluted to 25 ng/μl for the PCR. Three fluorescently labeled multiplex PCR assays were used to amplify exons from the FANCA and FANCC genes. The FANCC gene was used as an external control, since cell lines were classified as FA-A and no deletions have been described in extensive screening of the FANCC gene (Verlander et al. 1994; Gibson et al. 1996). Multiplex assay 1 amplified exons 10–12, 31, and 32 from the FANCA gene and exons 5 and 6 from the FANCC gene. Multiplex assay 2 amplified exons 5, 17, 35, and 43 from the FANCA gene and exons 5 and 6 from the FANCC gene. Multiplex assay 3 amplified exons 21 and 27 from the FANCA gene and exons 5 and 6 from the FANCC gene. All forward primers were labeled with either the fluorescent phosphoramidite 6-FAM or HEX dyes (PE Biosystems). Primer sequences, fluorescent dyes used, and sizes of products are shown in table 2. PCR amplifications were performed in 25 μl reactions with 125 ng DNA, 1 × Taq DNA polymerase buffer (as used for RT-PCR), and 200 μM each dNTP. Of each of the primer pairs, 0.2 μM was used for each of the three multiplexes, apart from 0.4 μM for FANCA exons 5, 11, 12, and 31. After an initial denaturation at 94°C for 3 min, 1.5 U Taq DNA polymerase (Promega) was added (hot start), and 18 PCR cycles of 93°C for 1 min, annealing for 1 min at either 60°C (multiplex 1 and 3) or 58°C (multiplex 2), and extension for 2 minutes at 72°C were performed, followed by a final extension for 5 min at 72°C. Only 18 PCR cycles were performed, to keep the reactions within the exponential phase of PCR (see Yau et al. 1996). An aliquot of the PCR product (4 μl) was added to 3.5 μl formamide loading buffer (95% formamide in 1 × TBE and 5 mg dextran blue/ml) and 0.5 μl internal lane size standard (Genescan-500 Rox; PE Biosystems). The samples were denatured for 5 min at 94°C and electrophoresed on a 5% denaturing polyacrylamide gel at 45 W for 6 h on an ABI 373 fluorescent DNA sequencer. Data were then analyzed by means of Genescan and Genotyper software, to obtain electrophoretograms for each sample. The position of the peaks indicates the size (in bp) of the exons amplified, and the areas under the peaks indicate the amount of fluorescence from the product. Figure 2 shows an example of the electrophoretograms obtained. On one gel, ⩽24 samples could be amplified and electrophoresed. At least four control DNAs were always included on each gel. The copy number of each exon amplified was established by importing the peak area values into an Excel spreadsheet and calculating a dosage quotient for each exon relative to all the other amplified exons in patients and controls (for an example see table 3). Peak areas from two control samples with approximately equal values were chosen, and the dosage quotient was calculated from them; values are typically within the range 0.77–1.25 (Yau et al. 1996). An average peak area of these controls was taken and compared with the peak area of the exon from the patient samples. As an example, in patient EUFA444 the dosage quotient for FANCA exon 10 and FANCC exon 5 is given by: DQ FANCA exon 10/FANCC exon 5 and is calculated by: [sample FANCA exon 10 peak area/sample FANCC exon 5 peak area] [control FANCA exon 10 peak area/control FANCC exon 5 peak area] = [1857/5301]/[6034/8180] = 0.47. After assessment of which exons were most frequently deleted from FANCA, multiplex 4 was developed (see table 2), with exon 1 of the myelin protein zero gene as the external control.
Table 2.
PCR Primer Sets for Multiplex Dosage Assays
| Exon | Primers (5′→3′) | Size (bp) |
| Multiplex 1: | ||
| FANCA: | ||
| Exon 10 | Forward, GAT TGT AGA AGT CTT GAT GGA TGT G | 259 |
| Reverse, ATT TGG CAG ACA CCT CCC TGC TGC | ||
| Exon 11 | Forward, GAT GAG CCT GAG CCA CAG TTT GTG | 301 |
| Reverse, AGA ATT CCT GGC ATC TCC AGT CAG | ||
| Exon 12 | Forward, CCA CAA CTT TTT GAT CTC TGA CTT G | 224 |
| Reverse, GTG CCG TCC ACG GCA GGC AGC ATG | ||
| Exon 31 | Forward, CAC ACT GTC AGA GAA GCA CAG CCA | 205 |
| Reverse, CAC GCG GCT TAA ATG AAG TGA ATG C | ||
| Exon 32 | Forward, CTT GCC CTG TCC ACT GTG GAG TCC | 369 |
| Reverse, CTC ACT ACA AAG AAC CTC TAG GAC | ||
| FANCC: | ||
| Exon 5 | Forward, CTG ATG TAA TCC TGT TTG CAG CGT G | 186 |
| Reverse, TCC TCT CAT AAC CAA ACT GAT ACA | ||
| Exon 6 | Forward, GTC CTT AAT TAT GCA TGG CTC TTA G | 293 |
| Reverse, CCA ACA CAC CAC AGC CTT CTA AG | ||
| Multiplex 2: | ||
| FANCA: | ||
| Exon 5 | Forward, ACC TGC CCG TTG TTA CTT TTA | 250 |
| Reverse, AGA ACA TTG CCT GGA ACA CTG | ||
| Exon 17 | Forward, CCC TCC ATG CCC ACT CCT CAC ACC | 207 |
| Reverse, AAA AGA AAC TGG ACC TTT GCA T | ||
| Exon 35 | Forward, GAT CCT CCT GTC AGC TTC CTG TGA G | 315 |
| Reverse, GCA TTT TCC CTG AGA TGG TAA CAC C | ||
| Exon 43 | Forward, GCC TGG CTG GCA ATA CAA CTC GAC | 223 |
| Reverse, GGC AGG TCC CGT CAG AAG AGA TGA G | ||
| FANCC: | ||
| Exon 5 | Forward, CTG ATG TAA TCC TGT TTG CAG CGT G | 186 |
| Reverse, TCC TCT CAT AAC CAA ACT GAT ACA | ||
| Exon 6 | Forward, GTC CTT AAT TAT GCA TGG CTC TTA G | 293 |
| Reverse, CCA ACA CAC CAC AGC CTT CTA AG | ||
| Multiplex 3: | ||
| FANCA: | ||
| Exon 21 | Forward, CAG GCT CAT ACT GTA CAC AG | 335 |
| Reverse, CAC CGG CTT GAG CTG GCA CAG | ||
| Exon 27 | Forward, CAG GCC ATC CAG TTC GGA ATG | 285 |
| Reverse, CCT TCC GGT CCG AAA GCT GC | ||
| FANCC: | ||
| Exon 5 | Forward, CTG ATG TAA TCC TGT TTG CAG CGT G | 186 |
| Reverse, TCC TCT CAT AAC CAA ACT GAT ACA | ||
| Exon 6 | Forward, GTC CTT AAT TAT GCA TGG CTC TTA G | 293 |
| Reverse, CCA ACA CAC CAC AGC CTT CTA AG | ||
| Multiplex 4: | ||
| FANCA: | ||
| Exon 5 | Forward, ACC TGC CCG TTG TTA CTT TTA | 250 |
| Reverse, AGA ACA TTG CCT GGA ACA CTG | ||
| Exon 11 | Forward, GAT GAG CCT GAG CCA CAG TTT GTG | 301 |
| Reverse, AGA ATT CCT GGC ATC TCC AGT CAG | ||
| Exon 17 | Forward, CCC TCC ATG CCC ACT CCT CAC ACC | 207 |
| Reverse, AAA AGA AAC TGG ACC TTT GCA T | ||
| Exon 21 | Forward, CAG GCT CAT ACT GTA CAC AG | 335 |
| Reverse, CAC CGG CTT GAG CTG GCA CAG | ||
| Exon 31 | Forward, CAC ACT GTC AGA GAA GCA CAG CCA | 268 |
| Reverse, CCC AAA GTT CTG GGA TTA CAG GCG TG | ||
| Myelin protein zero: | ||
| Exon 1 | Forward, CAG TGG ACA CAA AGC CCT CTG TGT A | 389 |
| Reverse, GAC ACC TGA GTC CCA AGA CTC CCA G |
Figure 2.
Electrophoretograms showing a deletion carrier of exons 10–12, detected with FANCA gene dosage multiplex 1
Table 3.
Statistical Profile of FANCA Dosage Multiplex 1
|
Peak Area in |
Dosage Quotient in |
|||
| Exon | Control | EUFA444a | FANCCExon 5 | FANCCExon 6 |
| Control: | ||||
| FANCC exon 5 | 8180 | 5301 | … | .99 |
| FANCC exon 6 | 4490 | 2949 | 1.01 | … |
| Test: | ||||
| FANCA exon 10 | 6034 | 1857 | .47 | .47 |
| FANCA exon 11 | 16967 | 5392 | .49 | .48 |
| FANCA exon 12 | 9068 | 3256 | .55 | .55 |
| FANCA exon 31 | 5466 | 3110 | .88 | .87 |
| FANCA exon 32 | 5838 | 3887 | 1.03 | 1.01 |
Heterozygous for a deletion of exons 10–12. For a sample calculation, see Methods subsection.
Results
A two-step mutation-screening strategy was devised to detect both small mutations—such as single-base substitutions—and heterozygous deletions of one or multiple exons of the FANCA gene. In the first step, the entire coding region was amplified by RT-PCR, and the fragments were sized on agarose gels. Aberrantly sized fragments were characterized by automated sequencing. The loss of an exon or the incorporation of intronic sequence led to the investigation of genomic DNA for splice-site mutations, whereas the loss of multiple exons revealed the presence of intragenic deletions. Full-length transcripts were screened by FCCM for nucleotide substitutions or small deletions and insertions. In the second step, genomic DNA was used to screen for deletions of one or more exons, by a fluorescent quantitative multiplex dosage assay that was based on an approach that we had originally developed to test for deletion and duplication carriers in Duchenne muscular dystrophy (Yau et al. 1996). In this assay, several exons of the FANCA gene were coamplified with two control exons from another locus, by use of fluorescently labeled primers, and the products were analyzed on an ABI 373 DNA-fragment analyzer. Heterozygous deletions were detected by a reduction in peak area of FANCA exons relative to those in controls. The two-step strategy was used to screen for mutations in RNA and DNA from 26 FA-A cell lines.
Aberrant FANCA Transcripts
RT-PCR analysis of the FANCA coding region in six overlapping fragments detected abnormally sized transcripts, in seven patients, that were found to be either splice-site mutations or intragenic deletions (all of the mutation data in this study are summarized in table 4). Patients EUFA123 and EUFA137 had reductions in size for fragments 2 and 4, respectively; sequencing of their genomic DNA revealed the splice-site mutations IVS6−2A→G and IVS16+3A→C. In patient EUFA779, fragment 4 contained an insertion of 90 bp of intron 15 sequence upstream of exon 16, and sequencing of genomic DNA around exon 16 from this patient revealed the splice-site mutation IVS15−1G→T. The intronic insertion resulted from the use of a cryptic splice-site in intron 15 and contained a stop codon (TGA) that would cause premature termination of translation. In patient EUFA755, fragment 4 showed no full-length product but did show two smaller products, which were revealed by sequencing to be two different deletions, of exons 15–17 and exons 16–20. PCR analysis of genomic DNA from this patient showed homozygous deletions of exons 16 and 17, which resulted from the overlap of the two intragenic deletions. In patients EUFA006 and EUFA417, reduced-size products in fragment 4 were shown, by sequencing, to be deletions of exons 15–20 and 16–17, respectively. Heterozygosity for deletions in this region was confirmed by quantitative PCR (see below). In patient EUFA854, a reduction in fragment 5 was a deletion of exons 29–31, again confirmed by quantitative PCR.
Table 4.
Mutations Detected in the FANCA Gene of FA Patients[Note]
| Patient | Mutation 1 | Effect | Exon(s) | Method of Detection | Mutation 2 | Effect | Exon(s) | Method of Detection |
| EUFA353 | 154C→Ta | R52X | 2 | FCCM | *5′ UTR–522*del | Deletion | *5′ UTR–5* | Dosage |
| BD237 | *5′ UTR–522*del | Deletion | *5′ UTR–5* | Dosage | 1115–1118deld | Frameshift | 13 | FCCM |
| EUFA1030 | *427–522*del | Deletion | *5* | Dosage | 987–990delb | Frameshift | 11 | Dosage |
| EUFA444 | *5′ UTR–1900*del | Deletion | *5′ UTR–21* | Dosage | 1115–1118deld | Frameshift | 13 | FCCM |
| EUFA750 | *5′ UTR–3066del | Deletion | *5′ UTR–31 | Dosage | 3788–3790dela,b | 1263delF | 38 | FCCM |
| BD812 | *5′ UTR–3066del | Deletion | *5′ UTR–31 | Dosage | Not detected | |||
| EUFA651 | 523–1359del | Deletion | 6–14 | Dosage | Not detected | |||
| EUFA123 | IVIS6−2A→Ga | Del13AA | I6 | RT-PCR | Not detected | |||
| EUFA186 | 597–1826del | Deletion | 7–20 | Dosage | Not detected | |||
| BD220 | 862G→T | E288X | 10 | FCCM | Homozygote | |||
| BD255 | 862G→T | E288X | 10 | FCCM | 4015delC | Frameshift | 41 | FCCM |
| EUFA056 | 1007–3066del | Deletion | 12–31 | Dosage | 1827–2778del | Deletion | 21–28 | Dosage/exon PCR |
| EUFA763 | 1191–1194dela | Frameshift | 13 | FCCM | 3971C→T | P1324L | 40 | FCCM |
| EUFA755 | 1360–1626del | Deletion | 15–17 | Dosage/RT-PCR | 1471–1826del | Deletion | 16–20 | Dosage/RT-PCR |
| EUFA006 | 1360–1826delc | Deletion | 15–20 | RT-PCR/Dosage | 3788–3790dela,b | 1263delF | 38 | FCCM |
| EUFA779 | IVS15−1G→T | Splicing | 16 | RT-PCR | Not detected | |||
| EUFA417 | 1471–1626deld,e | Deletion | 16–17 | RT-PCR/Dosage | 3788–3790dela,b | 1263delF | 38 | FCCM |
| EUFA137 | IVS16+3A→C | Exon skip | i16 | RT-PCR | Not detected | |||
| EUFA057 | 1771C→Tb | R591X | 19 | FCCM | Homozygote | |||
| EUFA134 | 1792G→Aa | D598N | 20 | FCCM | 1627–1900del | Deletion | 18–21 | Dosage |
| EUFA854 | 2779–3066del | Deletion | 29–31 | Dosage/RT-PCR | Not detected | |||
| EUFA583 | *2982–4365del | Deletion | *31–43 | Dosage | Not detected | |||
| BD32 | 3329A→C | H1110P | 33 | FCCM | Homozygote | |||
| EUFA413 | 3403–3405del | 1135delF | 34 | FCCM | Not detected | |||
| BD460 | 3629–3630insT | Frameshift | 37 | FCCM | Homozygote | |||
| EUFA097 | 3786C→G | F1262L | 38 | FCCM | 4080G→C | M1360I | 41 | FCCM |
Note.—An asterisk (*) denotes that the deletion endpoint is undefined.
Previously described by Wijker et al. (1999).
Previously described by Levran et al. (1997).
Previously described by Lo Ten Foe et al. (1996).
Previously described by Fanconi Anaemia/Breast Cancer Consortium (1996).
Previously described by Levran et al. (1998).
A high degree of alternative splicing of the FANCA gene was also detected. Exon 14 was commonly spliced out from fragment 3, and exon 30 from fragment 5. Both of these splicing events were seen in RNA prepared from lymphoblastoid control cell lines. An example of the shorter fragment produced in patients and controls, as a result of splicing out of exon 14, is shown in the electrophoretogram from the FCCM analysis in figure 1. Various other alternative splicing events were seen that, on sequencing, either produced mixed unreadable sequence or, by the detection of a mutation elsewhere in the gene, were excluded as the pathogenic mutation. For example, a transcript lacking exons 21–26 was seen in patient BD255, but this deletion was also present in controls, and both mutant alleles were found in other RT-PCR fragments from this patient.
FCCM Analysis
A total of 18 mutant alleles were detected by screening full-length transcripts from the six overlapping fragments of the FANCA coding region by FCCM, and these are listed in table 4. An example of the electrophoretic profiles produced by this analysis is shown in figure 1. Since PCR products are internally labeled by incorporation of fluorescent dUTP, the sizes of the two cleavage products accurately define the position of the mutation, which is then confirmed by sequencing of genomic DNA from the region. All 18 mutant alleles were detected by modification with hydroxylamine, which modifies only mismatched C bases. However, since both the probe and target were fluorescently labeled, and since biotinylated forward and reverse primers were used for each fragment, mutations such as A→G are detected as a mismatched C on the reverse strand. Although modification was also carried out with osmium tetroxide, no further mutations were found. The use of hydroxylamine alone has been shown to detect ⩽95% of all mutations (Rowley et al. 1995; Ellis et al. 1998). The procedure also detected numerous coding-sequence polymorphisms, including 1143G→T, 1235C→T, 1290G→A, 1501G→A, 1927C→G, 2151G→T, 2426G→A, and 4152G→C. These have been excluded as pathogenic mutations in other studies (Levran et al. 1997; Savino et al. 1997; Wijker et al. 1999; Fanconi Anemia Mutation Database) and have served as useful positive controls for the chemical cleavage.
Quantitative Fluorescent Multiplex PCR
Three separate quantitative multiplex PCR assays were established that collectively screened for heterozygous deletions in 11 exons across the FANCA gene. Exons were selected on the basis of robust amplification in multiplex PCR and by noting which exons were most frequently deleted as the work proceeded, and they included 5, 10–12, 17, 21, 27, 31, 32, 35, and 43. In these assays, the peak area of each exon is measured against all other exons in the multiplex, including two exons from a different locus, to produce a series of dosage quotients (for details, see Methods subsection). A typical electrophoretogram and a typical statistical profile of this analysis are shown in figure 2 and table 3, respectively. Genomic DNA from the 26 cell lines was used to screen for intragenic deletions by use of these three multiplex PCR reactions. A total of 17 large deletions were detected, which removed 1–31 exons of the FANCA gene (see table 4 and fig. 3). The multiplex assay also detects microdeletions within an exon, by means of size discrimination; in this way, patient EUFA1030 was found to be heterozygous for a 4-bp deletion in exon 11. The deletion data in figure 3 indicate that all deletions detected in the FA-A patients included one or more of the following five exons: 5, 11, 17, 21, and 31. A further multiplex was therefore designed that includes these five exons and exon 1 of the Myelin protein zero gene as an external control (see Methods subsection and table 2). This one multiplex, instead of the three other assays, can now be used for FANCA deletion screening.
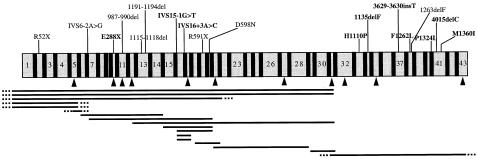
Figure 3.
Mutations detected in the FANCA gene in patients with FA. The figure is a schematic representation of the 43 exons of the FANCA gene. Novel mutations are shown in boldface. Large deletions are indicated by horizontal lines, which are dotted where endpoints are undefined. ▴ = Exons included in dosage multiplex assays.
RT-PCR analysis and polymorphic marker typing were used to map the approximate endpoints of the deletions. In six patients the most 5′ exon deleted was exon 5 (table 4). RT-PCR assays were therefore designed to amplify from exon 1 of the gene to a downstream exon known, from the dosage assay, to be present. These assays did not produce smaller transcripts, which suggested that the deletion endpoints extended upstream of the primer site in exon 1. A PCR was then carried out on genomic DNA, with a primer upstream of the FANCA 5′ UTR and a primer 3′ to the deletion, but no products were detected, again suggesting that the deletion extended beyond the promoter. The extent of these deletions was further investigated by typing the microsatellite polymorphic marker D16S3407, which is located 16 kb upstream of the 5′ UTR of the gene (Whitmore et al. 1998), in these patients and, if available, in parental samples. Since five of the six patients typed were homozygous for the marker (which has a heterozygosity of .77), the deletions may extend even farther upstream of the FANCA gene. This was confirmed in two of these five patients (EUFA353 and BD812), since parental DNA was available and informative with this marker, and both patients showed lack of inheritance of the parental allele that was linked to the deletion. The endpoints of deletions in EUFA cell lines 134, 186, 417, 651, 755, and 854 were defined by nested RT-PCR with primers outside the exons found to be deleted by the dosage assays followed by sequencing of the products (see table 4). The precise breakpoints of the exon 16–17 deletion in EUFA417 are different from the two deletions of these exons reported by Levran et al. (1998), since primers for those breakpoints did not generate the expected PCR products. In EUFA056, the endpoints of one deletion could be deduced as exon 21–28 since exons 21 and 28 were homozygously deleted and exons 20 and 29 were present; the other mutant allele in this patient is a deletion of exons 12–31, which was precisely defined in the dosage assay and overlaps the exon 21–28 deletion. The endpoint of the deletion in EUFA583 could not be defined, since the deletion extended beyond the 3′ UTR of the gene.
The analysis of FANCA deletion endpoints in this series of patients suggests at least four common breakpoints; there were four breaks in intron 31, four in intron 20, and up to five in the region 5′ to the gene (table 4). In view of the involvement of Alu-repeat elements in the generation of three published FANCA deletions (Centra et al. 1998; Levran et al. 1998), we searched the complete genomic sequence of the FANCA gene (AC005565, AC005567, and AC005360) for Alu repeats and compared the number of repeats with the number of known deletion breakpoints that we have detected in this and a previous study (Wijker et al. 1999). The results of this analysis (fig. 4) show that the most common breakpoints all occur in genomic regions that contain four or more Alu repeats and that none of the breakpoints occur in introns with fewer than two repeats. The presence of a deletion breakpoint correlated strongly with the presence of an Alu repeat (P<.001, Fisher's exact test), and linear regression analysis showed a highly significant correlation between the number of Alu repeats and the number of breakpoints detected (P=2.8×10-5).
Figure 4.
FANCA deletion breakpoints and Alu repeats. The number of deletion breakpoints includes those defined in this study and in the study by Wijker et al. (1999). The number of Alu-repeat elements shown for the 5′ end of the gene is for the sequence from the microsatellite D16S3407 to the initiation codon of FANCA. The number for the 3′ end is for 3 kb of sequence downstream of the stop codon.
FANCA Mutation Spectrum
A total of 33 different mutations were identified in this study, 23 of which were novel (fig. 3). This very heterogeneous spectrum included three splice-site mutations, two in-frame deletions of single–amino acid codons, three nonsense mutations, five missense mutations, five microdeletions or insertions that caused frameshifts, and 15 different large intragenic deletions (table 4 and fig. 3). The large intragenic deletions accounted for 17 (40%) of the 43 mutant alleles, making them by far the most common class of mutation in the FANCA gene. No single mutation was common, but E288X was found in two unrelated British patients, and 1263delF, which was originally described by Levran et al. (1997), was found in two French patients and one German patient. The pathogenic status of the five missense mutations was verified initially by confirming their cosegregation with the disease phenotype in the family and their absence in 100 ethnically matched control chromosomes. D598N is a nonconservative substitution that alters an amino acid residue that is conserved in the mouse Fanca gene (H. van de Vrugt, personal communication). H1110P is a nonconservative change that is not conserved in the mouse but has been shown to be nonfunctional in a cellular complementation assay (Kupfer et al. 1999). The other three (F1262L, P1324L, and M1360I) are conservative changes, but the altered residues are all conserved in the mouse. The sensitivity of the two-step mutation-screening assay can be deduced from the fact that at least one mutation was detected in all 26 FA-A cell lines tested, and 43 (83%) of the possible 52 mutant alleles were found.
Discussion
The modest detection rates reported from mutation screening of the FANCA gene, coupled with several reports of intragenic deletions (Centra et al. 1998; Levran et al. 1998; Wijker et al. 1999), led us to investigate whether these findings were connected. We therefore devised a mutation-screening protocol that would detect both small mutations, such as point mutations and microdeletions or insertions, and gross deletions that were present in the heterozygous state. The protocol was used to screen RNA and DNA from 26 cell lines that had been selected solely on the basis of their having been shown, by complementation analysis, to be FA group A. The quantitative PCR assay detected 17 deletions of the FANCA gene in these cell lines, which accounted for 40% of the mutant alleles observed. This result suggests that gross deletions of one or more exons of the FANCA gene are the most common form of mutation in this gene. The endpoints of 10 of the 17 deletions were established by RT-PCR and sequencing (table 4 and fig. 3). In six of the other deletions, genomic PCR with a primer from the FANCA 5′ UTR showed that the deletion is likely to extend upstream of the gene, and five patients were apparently homozygous for a highly informative microsatellite polymorphism, D16S3407, that lies 16 kb upstream of the initiation codon (Whitmore et al. 1998). This suggests that the deletion extends beyond this marker in at least some of these, as confirmed by family studies with this marker in two cases. In one of these cases, BD812, the deletion extends to exon 31 and must therefore remove ⩾80 kb of DNA. The remaining deletion extends 3′ to the last exon of FANCA, exon 43. The presence of several common breakpoints in this series of deletions and the evidence that three fully characterized deletions of FANCA appear to result from Alu-mediated recombination (Centra et al. 1998; Levran et al. 1998) led us to compare the breakpoints of all the deletions that we have detected in this and another study (Wijker et al. 1999) with the location of Alu repeats in FANCA that were obtained from its complete genomic sequence. This analysis (fig. 4) revealed a highly significant correlation between the breakpoints and the presence of multiple repeats and provides strong experimental support for the suggestion that this may be a common method for the generation of FANCA deletions (Centra et al. 1998; Levran et al. 1998). The frequency of these repeats in FANCA would then explain the high prevalence of gross deletions in its mutation spectrum. It might also indicate a higher mutation rate for FANCA as compared to other FA genes, which would explain why FA-A is by far the most common complementation group in this disorder (Joenje et al. 1997).
The two-step mutation-screening protocol that we developed in this study detected at least one mutation in all 26 cell lines screened and 43 (83%) of the expected 52 mutant alleles. This compares well with the results of other published studies. Savino et al. (1997) detected mutations in only 29% of Italian patients with FA, although complementation analysis showed that the prevalence of FA-A in Italy was >90% (Savoia et al. 1996). Levran et al. (1997) found mutations in 69% of patients unselected for complementation group, but 35% of these mutations were amino acid substitutions of unknown pathogenic status, and Wijker et al. (1999) detected mutations in 49% of probable FA-A patients. The highest detection rate was reported by Tachibana et al. (1999), who found mutations in 12 of 15 Japanese FA patients by direct sequencing; 22 (73%) of a possible 30 mutant alleles were detected. None of these studies included quantitative analysis of FANCA exon copy number. Our finding that 40% of the mutant alleles detected in 26 FA-A cell lines were heterozygous deletions of multiple exons suggests that mutation-detection rates can be substantially improved by incorporation of a quantitative assay into screening protocols. Since all of the mutations observed in our study involved one or more of exons 5, 11, 17, 21, and 31, we replaced the three multiplex PCRs that we developed initially by a single multiplex that incorporated these five exons and a control exon from an unrelated gene. This could serve as a useful and rapid initial molecular screen for FA in patients of unknown complementation group or of uncertain diagnosis.
The combination of FCCM and dosage analysis failed to detect 9 (17%) of a possible 52 mutant alleles. There are several possible explanations for this. The undetected mutations could be intragenic deletions of exons that were not included in the dosage multiplexes; collectively, the three multiplexes screened only 11 of the 43 exons, and the RT-PCR analysis would not detect all such deletions—for a variety of reasons, including the “noise” created by numerous alternative splicing events. The undetected mutations could also be complex genomic rearrangements, such as duplications or inversions, or they might conceivably be located in regions of the gene that were not screened in our protocol, such as the promoter or 3′ UTR. Finally, a small proportion of point mutations are not detected by FCCM if they are located in particular sequence contexts (Rowley et al. 1995; Ellis et al. 1998). Ultimately, no mutation-screening protocol is perfect, and a judgment has to be made as to whether a large amount of additional effort and expense can be justified in order to achieve 100% sensitivity of detection.
The very heterogeneous nature of the mutation spectrum in the FANCA gene is exemplified in this study: 33 different mutations were found in the 26 patients, of which 23 were novel mutations. The most interesting mutations, from the perspective of an attempt to describe the functional architecture of the FANCA protein, are those which do not cause premature termination of translation, such as the missense mutations and the deletions of single–amino acid codons. Almost all of these are clustered in the carboxy-terminal region of amino acid residues 1110–1360 (table 4), which may reflect the presence of an important functional domain involved in protein-protein interactions or in transport to the nucleus. In this group, only H1110P has, as yet, been confirmed as disrupting the function of the FANCA protein (Kupfer et al. 1999). Introduction of the others into the wild-type cDNA is in progress, in order to test them for absence of functional complementation.
Finally, the high sensitivity of the two-step screening procedure described in this article suggests that it may be usefully applied to mutation detection in other disorders. The utility of the multiplex quantitative assay for detection of deletions in the heterozygous state could be exploited to search for gross deletions in autosomal dominant disorders such as familial breast cancer, since evidence for such deletions in the BRCA1 gene is beginning to emerge (Puget et al. 1999). Also, in view of the general importance of Alu-mediated deletions in human genetic disorders (Purandare and Patel 1997) and the increasing availability of the complete genomic sequence of human genes, an initial electronic search for the location and density of such repeats may be a useful prelude to the design of mutation-screening strategies for a specific gene.
Acknowledgments
This work was supported by the European Union Biomed 2 program, the Fanconi Anemia Research Fund Inc. (USA), Fanconi Anemia Breakthrough (U.K.), and the Wellcome Trust. We thank the families with FA and the many clinicians associated with the European Fanconi Anemia Research Group (EUFAR), for providing blood samples; Dr. Cathryn Lewis, for the statistical analysis; and Michael Yau, for help with the gene dosage assay.
Electronic-Database Information
URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
- Fanconi Anemia Mutation Database, http://www.rockefeller.edu/fanconi/mutate
References
- Auerbach AD (1993) Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp Hematol 21:731–373 [PubMed]
- Centra M, Memeo E, d'Apolito M, Savino M, Ianzano L, Notarangelo A, Liu J, et al (1998) Fine exon-intron structure of the Fanconi anemia group A (FAA) gene and characterization of two genomic deletions. Genomics 51:463–467 [DOI] [PubMed]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159 [DOI] [PubMed]
- Cotton RGH, Rodrigues NR, Campbell RD (1988) Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci USA 85:4397–4401 [DOI] [PMC free article] [PubMed]
- De Winter JP, Waisfisz Q, Rooimans MA, van Berkel CGM, Bosnoyan-Collins L, Alon N, Bender O, et al (1998) The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat Genet 20:281–283 [DOI] [PubMed]
- Ellis TP, Humphrey KE, Smith MJ, Cotton RGH (1998) Chemical cleavage of mismatch: a new look at an established method. Hum Mutat 11:345–353 [DOI] [PubMed]
- Fanconi Anaemia/Breast Cancer Consortium (1996) Positional cloning of the Fanconi anaemia group A gene. Nat Genet 14:324–328 [DOI] [PubMed]
- Gibson RA, Morgan NV, Goldstein LH, Pearson IC, Kesterton IP, Foot NJ, Jansen S, et al (1996) Novel mutations and polymorphisms in the Fanconi anemia group C gene. Hum Mutat 8:140–148 [DOI] [PubMed]
- Ianzano L, D'Apolito M, Centra M, Savino M, Levran O, Auerbach AD, Cleton-Jansen AM, et al (1997) The genomic organization of the Fanconi anemia group A (FAA) gene. Genomics 41:309–314 [DOI] [PubMed]
- Joenje H, Lo Ten Foe JR, Oostra AB, van Berkel CG, Rooimans MA, Schroeder-Kurth T, Wegner RD, et al (1995) Classification of Fanconi anemia patients by complementation analysis: evidence for a fifth genetic subtype. Blood 86:2156–2160 [PubMed]
- Joenje H, Oostra AB, Wijker M, di Summa FM, van Berkel CG, Rooimans M, Ebell W, et al (1997) Evidence for at least eight Fanconi anemia genes. Am J Hum Genet 61:940–944 [DOI] [PMC free article] [PubMed]
- Kupfer G, Naf D, Garcia-Higuera I, Wasik J, Cheng A, Yamashita T, Tipping A, et al (1999) A patient-derived mutant form of the Fanconi anaemia protein, FANCA, is defective in nuclear accumulation. Exp Hematol 27:587–593 [DOI] [PubMed]
- Levran O, Doggett NA, Auerbach AD (1998) Identification of Alu-mediated deletions in the Fanconi anemia gene FAA. Hum Mutat 12:145–152 [DOI] [PubMed]
- Levran O, Erlich T, Magdolena N, Gregory JJ, Batish SD, Verlander PC, Auerbach AD (1997) Sequence variation in the Fanconi anemia gene FAA. Proc Natl Acad Sci USA 94:13051–13056 [DOI] [PMC free article] [PubMed]
- Lo Ten Foe JR, Rooimans MA, Bosnoyan-Collins L, Alon N, Wijker M, Parker L, Lightfoot J, et al (1996) Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nat Genet 14:320–323 [DOI] [PubMed]
- Puget N, Stoppa-Lyonnet D, Sinilnikova OM, Pages S, Lynch HT, Lenoir GM, Mazoyer S (1999) Screening for germ-line rearrangements and regulatory mutations in BRCA1 led to the identification of four new deletions. Cancer Res 59:455–461 [PubMed]
- Purandare SM, Patel PI (1997) Recombination hot spots and human disease. Genome Res 7:773–786 [DOI] [PubMed]
- Rowley G, Saad S, Giannelli F, Green PM (1995) Ultrarapid mutation detection by multiplex, solid-phase chemical cleavage. Genomics 30:574–582 [DOI] [PubMed]
- Savino M, Ianzano L, Strippoli P, Ramenghi U, Arslanian A, Bagnara GP, Joenje H, et al (1997) Mutations of the Fanconi anemia group A gene (FAA) in Italian patients. Am J Hum Genet 61:1246–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoia A, Zatterale A, Del Principe D Joenje H (1996) Fanconi anaemia in Italy: high prevalence of complementation clusters. Hum Genet 97:599–603 [DOI] [PubMed]
- Strathdee CA, Duncan AM, Buchwald M (1992) Evidence for at least four Fanconi anaemia genes including FACC on chromosome 9. Nat Genet 1:196–198 [DOI] [PubMed]
- Tachibana A, Kato T, Ejima Y, Yamada T, Shimizu T, Yang L, Tsunematsu Y, et al (1999) The FANCA gene in Japanese Fanconi anemia: reports of eight novel mutations and analysis of sequence variability. Hum Mutat 13:237–244 [DOI] [PubMed]
- Verlander PC, Lin JD, Udono MU, Zhang Q, Gibson RA, Mathew CG, Auerbach AD (1994) Mutation analysis of the Fanconi anemia gene FACC. Am J Hum Genet 54:595–601 [PMC free article] [PubMed]
- Waisfisz Q, Saar K, Morgan NV, Altay C, Leegwater PA, de Winter JP, Komatsu K, et al (1999) The Fanconi anaemia group E gene, FANCE, maps to chromosome 6p. Am J Hum Genet 64:1400–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore S, Crawford J, Apostolou S, Eyre H, Baker E, Lower KM, Settasatian C, et al (1998) Construction of a high-resolution physical and transcription map of chromosome 16q24.3: a region of frequent loss of heterozygosity in sporadic breast cancer. Genomics 50:1–8 [DOI] [PubMed]
- Whitney M, Thayer M, Reifsteck C, Olson S, Smith L, Jakobs PM, Leach R, et al (1995) Microcell mediated chromosome transfer maps the Fanconi anaemia group D gene to chromosome 3p. Nat Genet 11:341–343 [DOI] [PubMed]
- Wijker M, Morgan NV, Herterich S, van Berkel CGM, Tipping AJ, Schindler D, Gille JJP, et al (1999) Heterogeneous spectrum of mutations in the Fanconi anaemia group A gene. Eur J Hum Genet 7:52–59 [DOI] [PubMed]
- Yau SC, Bobrow M, Mathew CG, Abbs SJ (1996) Accurate diagnosis of carriers of deletions and duplications in Duchenne/Becker muscular dystrophy by fluorescent dosage analysis. J Med Genet 33:550–558 [DOI] [PMC free article] [PubMed]