SUMMARY
Alternative splicing is prevalent in the mammalian brain. To interrogate the functional role of alternative splicing in neural development, we analyzed purified neural progenitor cells (NPCs) and neurons from developing cerebral cortices, revealing hundreds of differentially spliced exons that preferentially alter key protein domains—especially in cytoskeletal proteins—and can harbor disease-causing mutations. We show that Ptbp1 and Rbfox proteins antagonistically govern the NPC-to-neuron transition by regulating neuron-specific exons. While Ptbp1 maintains apical progenitors partly through suppressing a poison exon of Flna in NPCs, Rbfox proteins promote neuronal differentiation by switching Ninein from a centrosomal splice form in NPCs to a non-centrosomal isoform in neurons. We further uncover an intronic human mutation within a PTBP1 binding site that disrupts normal skipping of the FLNA poison exon in NPCs and causes a brain-specific malformation. Our study indicates that dynamic control of alternative splicing governs cell fate in cerebral cortical development.
Keywords: Filamin A, Ninein, Ptbp1, Rbfox, microcephaly, periventricular nodular heterotopia, mother centriole
INTRODUCTION
The neocortex is phylogenetically the newest part of the human brain with its projection neurons originating from waves of neurogenesis initiated by apical radial glial cells (aRG), followed by extensive radial neuronal migration (Gao et al., 2014; Greig et al., 2013; Huttner et al., 2013; Lui et al., 2011). RGCs maintain long basal processes attached to the pial membrane and undergo mitosis near their apical processes in the ventricular zone (VZ) or outer subventricular zone (oSVZ) to self renew or produce other types of progenitors including intermediate progenitors (IPs). IPs undergo limited numbers of divisions to produce neurons. In humans, the massive expansion of the cortex is accompanied by additional numbers of cell divisions, progenitor types and neuronal circuits by mechanisms that are poorly understood (Lui et al., 2011).
Alternative splicing (AS) regulates over 90% of multi-exon protein-coding genes in humans and exerts an evolutionarily conserved posttranscriptional control on genome-wide and tissue-specific gene expression (Barbosa-Morais et al., 2012; Merkin et al., 2012; Wang et al., 2008). Transcriptional profiling of mammalian forebrain has revealed dynamic AS changes between different brain regions (Johnson et al., 2009), cortical layers (Belgard et al., 2011) or developmental stages (Dillman et al., 2013; Yan et al., 2015). Dysregulation of AS in human brain by RBFOX1 mutations or disturbed nSR100 levels has been associated with intellectual disability and autism spectrum disorders (ASD) (Bhalla et al., 2004; Irimia et al., 2014; Sebat et al., 2007). nSR100, Ptbp1 and Rbfox proteins have also been reported to regulate AS of neuronal microexons (Irimia et al., 2014; Li et al., 2015). Recent studies have generated an unprecedented view of AS in cortical development (Darnell, 2013; Li et al., 2007; Raj and Blencowe, 2015; Vuong et al., 2016), but the physiological impact of alternative splicing on cortical progenitor and neuronal fates remains unclear. On the one hand, systematic studies of cortical NPCs concentrated mostly on gene-level instead of exon-level expression or detected rather subtle AS changes (Ayoub et al., 2011). On the other hand, previous AS studies of the developing cerebral cortex centered on either RNA binding proteins (RBP) or individual alternative exons, rather than taking a global view of cell type-specific regulation. Direct comparative investigations of alternative exon usage between cortical NPCs and neurons in a physiological context are not yet available.
We performed unbiased RNA sequencing (RNA-Seq) comparison of NPCs and neurons isolated directly from developing mouse and human cerebral cortices, and identified extensive and conserved AS switches during cortical NPC differentiation. We found that alternative splicing preferentially regulates genes encoding cytoskeleton proteins, modulates protein subcellular localization, and involves genes essential for brain development in mice and/or humans. Our results on dynamic switching of Ninein and Filamin A isoforms by Rbfox1/2/3 and Ptbp1 proteins reveal developmental roles of alternative splicing in regulating centriolar dynamics, NPC self-renewal and differentiation, uncovering widespread functions of alternative splicing in cerebral cortical development.
RESULTS
RNA Sequencing of Sorted NPCs and Neurons from Developing Mouse Cerebral Cortex Uncovers Extensive Alternative Exon Usage during Cortical NPC Differentiation
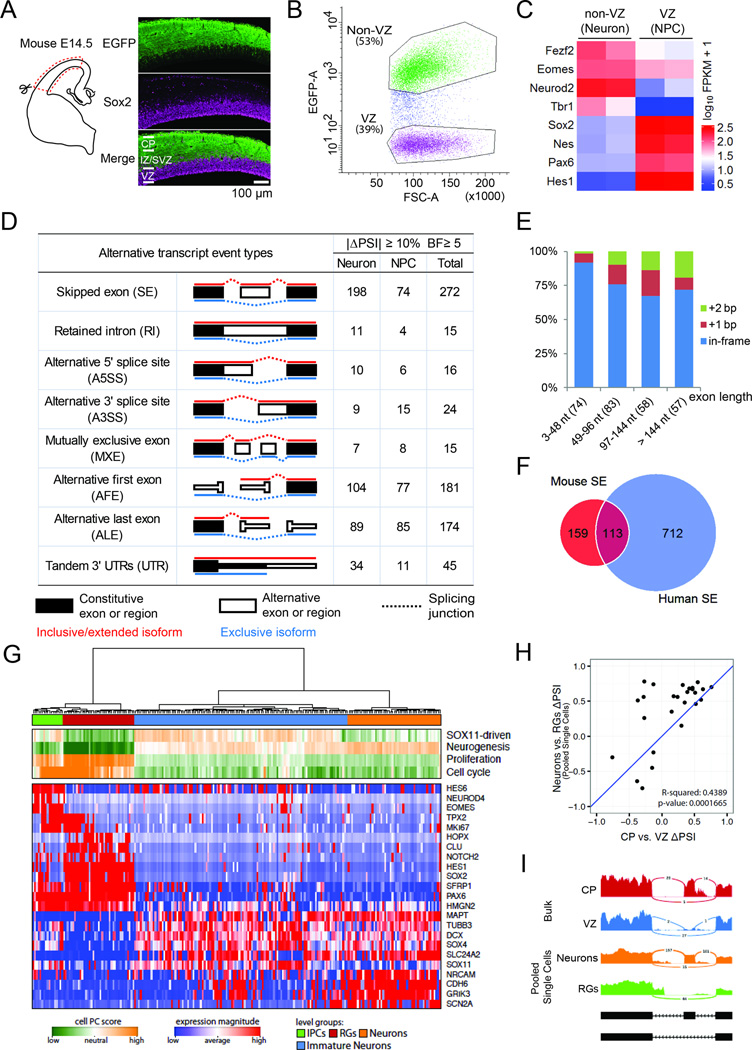
Using a Tbr2-EGFP transgene driving EGFP in dorsal cortex (Figure S1A) (Gong et al., 2003), we found that E14.5 VZ NPCs (Sox2+; EGFP−) are well separated from IPs in the subventricular zone (SVZ, Tbr2+; EGFP+) and differentiating neurons in the intermediate zone (IZ) and cortical plate (CP, Sox2-; EGFP+, Figure 1A and Figure S1A). We isolated VZ NPCs (EGFP−) and non-VZ cells (EGFP+) from E14.5 Tbr2-EGFP cerebral cortex (Figure 1B), and confirmed their identities: (1) strand-specific RNA-Seq and quantitative PCR of sorted cells showed that NPC genes Sox2, Pax6, Nes and Hes1 were highly enriched in the (EGFP−) cells while Tbr1, Fezf2 and Neurod2 were enriched in the (EGFP+) cells (Figure 1C, Figure S1B); (2) the fourth exon of REST, expressed in differentiating neurons (Raj et al., 2011), was depleted from sorted (EGFP−) cells (Figure S1C); (3) 93.5% of sorted (EGFP−) cells were Sox2 positive (Figure S1D–E); (4) gene ontology analysis revealed that cell cycle, chromosomal and DNA metabolic genes were enriched in (EGFP−) cells, while neuron differentiation and projection genes were enriched in (EGFP+) cells (Figure S1F). These results indicate that we successfully isolated and analyzed VZ NPCs (EGFP−) and a mixture of IPs and neurons outside the VZ (hereafter referred to as non-VZ or neuron) from developing mouse cerebral cortex.
Figure 1. Extensive and Conserved Alternative Exon Usages During Cerebral Cortical NPC Differentiation.
A) Immunostaining of E14.5 Tbr2-EGFP mouse dorsal cerebral cortex with anti-EGFP (green) and anti-Sox2 (magenta).
B) Fluorescence-activated cell sorting (FACS) of green and non-green cells from E14.5 Tbr2-EGFP mouse dorsal cerebral cortex.
C) Heatmap of RNA-Seq results showing differential gene expression between sorted VZ and non-VZ cells.
D) Number of alternative exons between neural progenitor cells (NPC) and differentiating neurons.
E) Histogram showing the size of mouse SEs (x axis) and the percentages of SEs that cause an in-frame insertion or a frame shift (y axis).
F) Conserved SEs in mouse and human cortical neurogenesis.
G) PAGODA analysis distinguishes 224 single fetal cortical cells (Camp et al., 2015) into 4 clusters (green, blue, red, yellow). Shown at the bottom are gene expression patterns for selected marker genes.
H) ΔPSI scores between pooled single cells and bulk samples show significant correlation.
I) Sashimi plots from bulk and single cell analyses showing the last three exons of CERS5.
See also Figure S1.
We compared alternative exon usage between E14.5 VZ NPCs and non-VZ cells using the mixture-of-isoforms (MISO) statistical model, which assigned a ‘percentage spliced in’ (PSI) value to each exon by estimating its abundance compared to adjacent exons (Katz et al., 2010). We found that 622 exons were differentially spliced between mouse NPCs and neurons (|ΔPSI|≥10% and Bayes factor ≥5, the same criteria used hereafter if not specified), with 345 showing higher inclusion in neurons and 277 higher in NPCs. We analyzed VZ and CP samples from two additional RNA-Seq datasets (Ayoub et al., 2011; Fietz et al., 2012), and found 742 AS changes shared by at least two of the three datasets (Figure 1D, Figure S1G). 272 cassette exons or skipped exons (SE) comprised the largest portion of AS events, with 198 (73%) SEs showing higher inclusion in neurons than in NPCs (Figure 1D), 255 (94%) SEs shorter than 300 nt (Figure S1H), and 61 (22%) SEs causing frame shifts (Figure 1E). These results indicate that hundreds of alternative exon usages occur when VZ NPCs differentiate in mice.
Extensive Alternative Exon Usage during NPC Differentiation in the Developing Human Cerebral Cortex
The developing human cortex has an enormously expanded oSVZ housing the largest population of dividing progenitors, the most prominent being outer radial glial (oRG) cells that resemble aRG cells but lack apical processes. We compared laser microdissected oSVZ to VZ, inner SVZ (iSVZ) and CP (Fietz et al., 2012), as well as RNA-seq data from purified aRG and oRG cells (Johnson et al., 2015), and found that most splicing switches occur between neural progenitors in the VZ and neurons in the CP (2582 exons, Figure S1I–S1J), with 31% of human cassette exons (|ΔPSI|≥15%, Bayes factor ≥10) causing a translational frame shift (Figure S1K). When SE, A3SS, A5SS, MXE and RI (Figure 1D) were considered, 38% of mouse differential splicing events were also differentially spliced in human, including 113/272 (42%) mouse SEs (Figure 1F).
To compare AS between pure NPC and neuronal populations, we further analyzed RNA-Seq data of 224 single cells from three human fetal cortices (Camp et al., 2015). Using the unbiased pathway and gene set over dispersion analysis approach (PAGODA) (Fan et al., 2016), we were able to identify distinct transcriptional subpopulations corresponding to RGs, IPs, immature neurons, and neurons (Fig 1G). We then pooled single RGs and single neurons in silico and extended our MISO analysis to these putatively pure RG and mature neuron populations, which gave rise to 298 significant cassette exon changes. Consistent with bulk samples, more SEs show higher inclusion in neurons (166) than in RGs (132). For 26 SEs that were also identified in human bulk samples, ΔPSIs are significantly correlated between bulk and pooled single cell analyses (r^2 = 0.44, p-value = 1.7e-4, Fig 1H–1I). These results indicate that extensive alternative exon usages occur when VZ NPCs differentiate in mouse and human cerebral cortex and many of them may critically regulate gene function by shifting translational frames, though we focus here mainly on those shared by mouse and human to allow functional analysis of their mechanism.
Cytoskeleton Genes Are Preferentially Regulated by Alternative Splicing during Neurogenesis
Gene ontology analysis for differentially spliced exons revealed that cytoskeleton genes are overrepresented in mouse and human (Figure 2A, Figure S2A–S2B). KEGG pathway analyses of alternative exons also revealed enriched functions in regulating tight junctions and actin cytoskeleton (Figure S2C and data not shown). Differential mouse SEs between E14.5 NPCs and neurons were validated by RT-PCR, and many of these events were switch-like (|ΔPSI| > 50%, Figures 2B–2F, and S2D–S2E, Table S1). Notably, alternatively spliced cytoskeleton genes encoded functionally interconnected protein networks (Figure 2G–2I) involved in NPC proliferation, neuronal migration and neuronal differentiation. Over a dozen genes alternatively spliced during the NPC-to-neuron transition--including Add1, Ank2, Flna, Kif2a, Macf1, Scrib, and Syne2--have previously been shown to be required for normal cerebral cortical development in mice (Table S2), and certain alternative exons harbor mutations that associate with human brain disorders such as microcephaly and autism (Figure 2J–2M and below), suggesting cell type-specific mechanisms of human diseases.
Figure 2. Alternative Splicing Preferentially Regulates Genes Encoding Cytoskeleton Proteins.
A) Ontology analysis of genes that are alternatively spliced between E14.5 mouse NPCs and differentiating neurons, showing the top 10 ranked terms and count of genes (in parenthesis).
B) RT-PCR analyses validate 57 AS events identified by MISO. ΔPSI = PSI (Neuron) – PSI (NPC).
C) to F) RT-PCR validation of alternatively spliced SEs related to microtubule C), actin (D), Add1-Ank2-Epb4.1 complex (E) and synapse (F). “E14.5 cc” represents unsorted E14.5 mouse cerebral cortex.
G) to I) Cartoon illustrations of alternatively spliced genes involved in NPC proliferation (G), radial neuronal migration (H) and neuronal differentiation (I).
J) RNA-Seq reads of human NIN gene in GW13-16 VZ and CP (left), showing that the 2139 bp alternative exon is included in VZ (shaded in light blue). Red bars indicate mutations associated with microcephaly, one of which lies in the AS exon (Dauber et al., 2012). The black arrows here and in all following genome browser figures indicate the direction of gene transcription.
K) qRT-PCR results show that the 2121 bp mouse homologous Nin exon is included in VZ but skipped in non-VZ cells. Data are represented as mean +/− SEM.
L) RNA-Seq reads of human ANK2 gene showing the 6255bp alternative exon (shaded) is included in CP. Red bars indicate mutations associated with autism spectrum disorder, two of which lie in the AS exon (Iossifov et al., 2014).
M) qRT-PCR analysis showing that the mouse homologous Ank2 exon of L) is included in non-VZ and skipped in VZ. Data are represented as mean +/− SEM.
Alternatively Spliced Exons Alter Protein Domains and Subcellular Localization
Exons that are differentially spliced between NPCs and neurons are remarkable in the extent to which they critically involve essential protein domains. Among 61 splicing changes in 49 genes regulating cytoskeleton (Table S3), 20 (33%) of them caused insertion or deletion of one or more entire protein domains (Figure 3A–3B, Figure S3A–S3C). Multiple genes showed AS of membrane-targeting domains (Figure 3B), which typically regulate subcellular localization. Another 13 (21%) alternative exons--including a microexon--inserted extra amino acids (extraAA) into well-defined protein domains (Figure 3A, Figure S3A and S3C). Thus, more than half (54%) of differentially spliced regions directly regulate protein domains.
Figure 3. Cell Type-specific Alternative Splicing Translocates Ninein from Centrosome in NPCs to Non-centrosomal Loci in Neurons.
A) Impact of 61 cytoskeleton-related and conserved AS events on modular protein domains. AA, amino acids. CT, C-terminus.
B) Alternative splicing alters protein domains that regulate subcellular localization. Gene names are followed by ΔPSI values. Differential protein domains are shaded red (neuron) or cyan (NPC).
C) RNA-Seq reads in E14.5 mouse VZ and CP, and GW13-16 human VZ and CP showing that the alternative Nin (NIN) exon 29 is specifically included in neurons (CP).
D) RT-PCR validates the specific inclusion of Nin exon 29 in differentiating neurons.
E) Alignment of Nin exon 30, showing that insertion of exon 29 introduces a conserved premature stop codon.
F) –G) Cartoon illustration of EGFP-Nin fusion constructs and their subcellular localization in transfected cells.
H) Left: co-IP of transfected Ninein isoforms showing that Nin-NPC, but not the Nin-Neuron isoform, pulls down CEP170 and CEP250. Right: co-IP showing that endogenous Ninein interact with CEP170 and CEP250 in Hela cells.
I) siRNA knockdown of CEP250 disrupts the centrosomal localization of Ninein.
Alternative Splicing Switches Ninein from Centrosome in NPCs to Non-centrosomal Loci in Neurons
We have shown a large Nin alternative exon 18 (> 2000 nt) that is almost exclusively included in mouse and human NPCs but skipped in neurons (Figure 2J–2K, ΔPSI = −83.5%). In contrast, we also identified a conserved 61 nt exon (exon 29) of Nin that is specifically included in neurons (CP) but skipped in NPCs (Figure 3C–3D, ΔPSI = 74.0%), introducing a frame shift and truncating the C-terminus of Nin (Figure 3E–3F). Nin associates with the mother centriole to anchor microtubules, with its C-terminal segment targeting efficiently to centrosomes (Delgehyr et al., 2005). Although Nin translocates from centrosome to noncentrosomal loci during brain progenitor cell differentiation, the underlying mechanism remains unknown (Baird et al., 2004; Ohama and Hayashi, 2009).
We examined the effects of exon 18 and exon 29 on Nin subcellular localization and found that the C-terminal centrosome-localization signal of Nin (Nin-NPC-CT) is lost with the inclusion of alternative exon 29, causing the neuronal Nin isoform (Nin-Neuron) to be diffusely localized in cytoplasm when highly expressed (Figure 3F–3G), associating with microtubules at non-centrosomal loci (Figure S3D–S3F). Targeted co-immunoprecipitation (co-IP) screen of centrosomal proteins allowed us to uncover that the Nin-NPC-CT specifically pulled down CEP250 (Figure 3H), and CEP250 knockdown disrupted the centrosomal localization of Nin (Figure 3I, Figure S3G). We further found that CEP170 preferentially interacts with Nin-NPC through exon 18, but not the Nin-Neuron isoform (Figure 3H, Figure S3I). Given that CEP170 relies on Nin for its centrosomal localization (Graser et al., 2007), our data suggest that the Nin-CEP170 interaction is mediated by Nin exon18 and is dynamically regulated by alternative splicing. These results indicate a two-fold alternative splicing mechanism: (1) inclusion of neuronal exon 29 dissociates Nin from CEP250 and centrosome, causing Nin to adopt a non-centrosomal localization in neurons; and (2) skipping of Nin exon 18 further dissociates CEP170 from Nin in neurons.
Endogenous Nin-Neuron is significantly elevated in postmitotic neurons during cortical neurogenesis (Figures S4A–S4B). Ectopic expression of Nin-Neuron, but not Nin-NPC, led to significant neural progenitor depletion from the VZ (Figure 4A–4B) and defective neuronal migration into the CP (Figure S4C). We performed pair-cell assays, and found Nin-Neuron expression causes significantly more NPCs to become neurons compared to control or the Nin-NPC isoform (Figure 4C–4D). Expression of Nin-Neuron disperses endogenous Nin and Dctn1--a key dynactin subunit mediating cytoplasmic dynein-cargo binding--from the centrosome (Figure 4E–4F), suggesting a dominant negative mechanism in regulating cell fate. We thus created a NIN knockout cell line (Chr14:51,259,547delT (hg19), p.Arg107Thrfs*16, Figure S4D) and found that loss of centrosomal NIN disrupted normal mitotic cleavage planes (Figure 4G–4H). These results indicate that endogenous switching of Nin-NPC to the Nin-Neuron isoform is sufficient to convert cortical NPCs to neurons, suggesting that neurogenesis is normally controlled at least in part by alternative splicing of Nin.
Figure 4. Ninein Neuronal Isoform Promotes NPC Differentiation.
A–B) Expressing Nin-Neuron, but not the Nin-NPC isoform, in E13.5 mouse brains leads to fewer NPCs in the VZ at E15.5. Numbers in parentheses indicate the number of embryos (n) analyzed. Data are represented as mean +/− SD.
C–D) Pair-cell analyses showing that expression of Nin-Neuron promotes neuron production. P, progenitor; N, neuron. Data are represented as mean +/− SD.
E) Expression of Nin-Neuron isoform decreases the level of endogenous NIN at the centrosome.
F) Expression of Nin-Neuron disperses Dctn1 away from centrosome.
G) NIN signal is diminished in NIN knockout cells.
H) NIN loss-of-function leads to defective mitotic spindles. Data are represented as mean +/− SD.
See also Figure S4.
Aberrant Splicing of A Cryptic Poison Exon Causes A Unique Brain Malformation
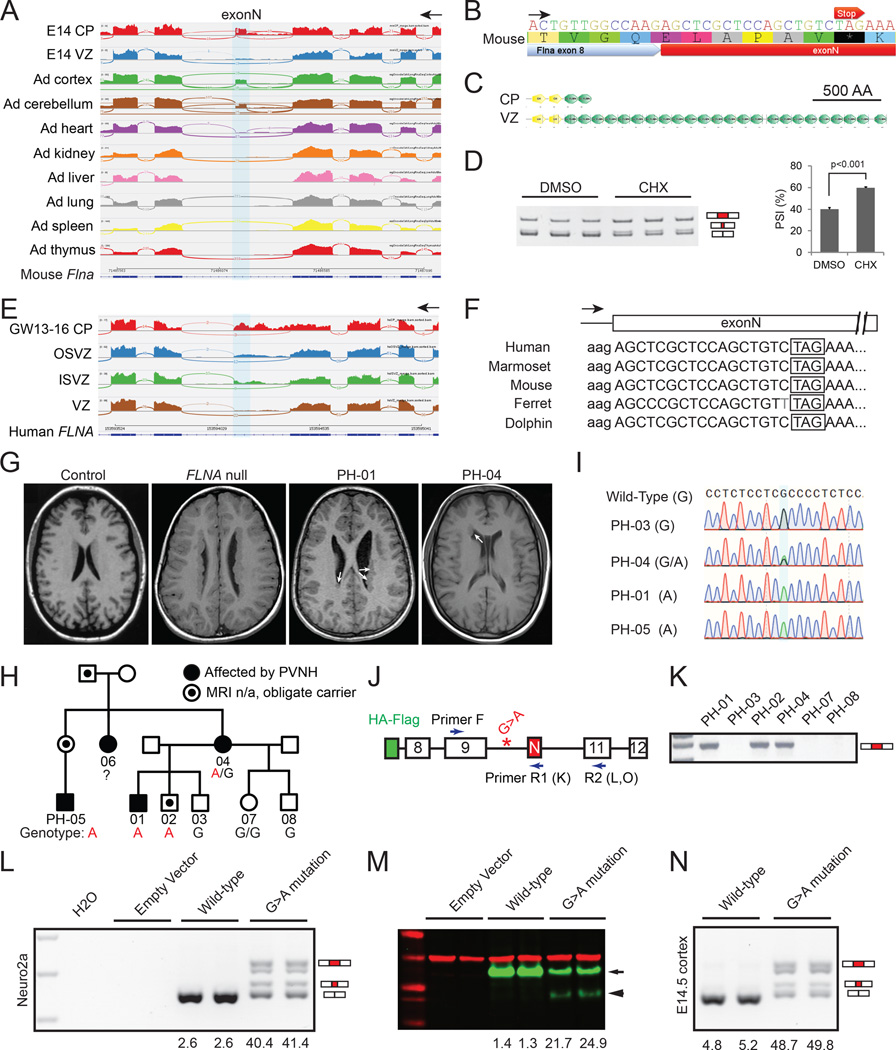
Splicing analysis identified a highly conserved but not previously annotated 57 nt exon in Flna that is skipped in E14.5 NPCs (VZ) and most adult tissues, but included in mRNAs from E14.5 neurons (CP) as well as adult cortex and cerebellum (Figure 5A). Surprisingly, an in-frame stop codon was embedded in this neuron-specific Flna exon (Figure 5B and Figure S5A), inclusion of which causes early truncation of Flna protein (482 of 2647 amino acids, Figure 5C), and/or nonsense-mediated mRNA decay (NMD, Figure 5D). We thus name the Flna SE “exonN” (N stands for Null in Neuron), and found that the human homologous exonN is skipped in VZ while included in CP (Figure 5E). The skipping of exonN in cortical NPCs creates a full-length protein, whereas splicing into this exon in neurons would give a truncated protein or unstable mRNA. Remarkably, genomic sequences of exonN including the embedded stop codon are conserved across multiple placental mammalian species, though it is not observed outside mammals (Figure 5F).
Figure 5. Cell Type-specific Alternative Splicing of Filamin A in Cerebral Cortical Development and Human PVNH.
A) RNA-Seq reads around Flna exonN showing its higher inclusion in adult cerebral cortex, cerebellum and E14.5 CP, in comparison to E14.5 VZ and other non-neural adult tissues.
B) Flna exonN has an in-frame stop codon.
C) ExonN inclusion is predicted to truncate Flna protein.
D) Blocking of NMD by cycloheximide (CHX) in primary hippocampal neurons cultured in vitro for 1 day increases inclusion of exonN. Data are represented as mean +/− SEM.
E) RNA-Seq reads showing the inclusion of exonN and retention of its upstream intron in fetal human CP.
F) Filamin A exonN is conserved across representative placental mammals.
G) T1 brain MRI shows PVNH (white arrows) in affected individuals PH-01 (male) and PH-04 (female), compared to a healthy control and an unrelated individual with a FLNA null mutation.
H) Pedigree showing inheritance of PVNH and the rare c.1429 +182 G>A mutation.
I) Sanger sequencing traces showing the rare FLNA variant c.1429+182 G>A.
J) A cartoon illustrating the FLNA mini-gene construct and primers used for RT-PCR. N-terminal HA and Flag tags were fused in-frame with exon 8.
K) RT-PCR using primer F and R1 (inside exonN) detecting the abnormal exonN inclusion in blood samples of affected individuals.
L) RT-PCR (primer pair F-R2) results on FLNA mini-genes expressed in Neuro2a cells, showing that the c.1429+182 G>A mutation caused > 40% of FLNA transcripts to include exonN.
M) Western blot of transfected Neuro2a cells detecting the WT mini-FLNA (green, arrow) and the truncated form (arrow head). Red, anti-Gapdh.
N) RT-PCR (primer pair F-R2) results of E14.5 mouse brains electroporated FLNA mini-genes on E13.5, showing that the G>A mutation caused abnormal inclusion of exonN.
See also Figure S5.
The presence of poison exons encoding translational stop codons reported here and by other groups (Yan et al., 2015) suggests a potential mechanism of human disease that to our knowledge has not been previously described: mutations that disrupt normal suppression of a poison exon would inactivate protein expression in a cell that normally skips the exon. We tested this model by studying FLNA, since heterozygous null mutations in females typically cause periventricular heterotopia (PVNH), where neurons form nodules along the VZ, reflecting failure of normal migration (Fox et al., 1998). We hypothesized that, since abnormal inclusion of exonN will create a FLNA null allele (Figure 5B–5F), mutations that promote exonN inclusion in NPCs may lead to PVNH.
Sequencing FLNA exonN and its flanking introns in 221 individuals with genetically unexplained PVNH revealed a rare variant FLNA-as (ChrX: 153,594,210 C>T, c.1429+182 G>A) in a large family in which multiple individuals show PVNH that is notably milder than that caused by FLNA null alleles (Figure 5G–5I). Unlike typical females with heterozygous FLNA null mutations, who have abnormally located neurons lining the entire lateral ventricles bilaterally, the PVNH in FLNA-as patients is striking, but limited to smaller segments of the VZ, often asymmetrically (Figure 5G). The FLNA-as mutation lies in the upstream intron of exonN (Figure 5J), and leads to abnormal exonN inclusion in patient blood cells, where exonN is normally skipped (Figure 5K). Furthermore, FLNA null mutations are usually male lethal, yet affected males in this family do not show early lethality. Males and females with null FLNA mutations also show variably penetrant congenital heart disease and severe vascular catastrophes (Fox et al., 1998), but the FLNA-as pedigree shows no clear evidence of cardiovascular disease, suggesting that the FLNA-as mutation may cause a central nervous system (CNS)-specific and cell type-specific phenotype.
We expressed wild-type (WT) and mutated FLNA mini-genes in Neuro2a cells (Figure 5J and 5L–5M) and in E14.5 mouse cortical NPCs (Figure 5N) and found that the WT mini-gene produced transcripts that skipped exonN (PSI<5%), while the mini-gene with the FLNA-as mutation produced 40%–50% of FLNA transcripts that abnormally included exonN or the last 8-nt of exonN that creates a frame shift (Figure 5L, 5N, Figure S5B). FLNA-as mutation led to a truncated FLNA mini-protein and reduced levels of mini-proteins (Figure 5M). These results confirm that the c.1429+182 G>A mutation promoted abnormal exonN inclusion in NPCs, creating a cell type- and tissue-specific FLNA partial loss-of-function and an atypical PVNH syndrome.
Ptbp1 and Rbfox1/2/3 Antagonistically Regulate Neuronal Exon Inclusion in the Developing Cerebral Cortex
We used DREME (Bailey, 2011) to identify discriminative cis-regulatory sequence motifs enriched in the flanking introns of cassette exons that are preferentially included in neurons. The only two significant motifs identified correspond to binding motifs of Ptbp1/2 (CU(C/U)UCUU, found within 200 nts in the upstream intron, p-value=3.6e-9), and Rbfox1/2/3 (GCAU(G/A), within 200 nts in the downstream intron, p-value =1.3e-8, Figure 6A). Notably, 61% of alternatively spliced neuronal exons had Ptbp1/2 and/or Rbfox1/2/3 regulatory motifs (Figure S6A). Analyses of Rbfox HITS-CLIP and Ptbp1 iCLIP datasets from neuronal samples (Linares et al., 2015; Weyn-Vanhentenryck et al., 2014) further support that many cortically regulated exons exhibit direct binding by these proteins (Figure S6B, Table S4 and below).
Figure 6. Alternative Exons Showing Higher Inclusion in Neurons Are Antagonistically Regulated by Ptbp1 and Rbfox1/2/3 Proteins.
A) Unbiased motif analysis of alternative exons with higher inclusion in differentiating neurons reveals the enrichment of CU(C/U)UCUU in the 200 nt 5’ upstream region and GCAU(G/A) in the 200 nt downtream intronic region.
B) RNA-Seq results of sorted E14.5 mouse cortical cells showing that Ptbp1 is enriched in NPCs (VZ) and Rbfox1/2/3 transcripts are enriched in non-VZ cells. Data are represented as mean +/− SEM.
C) A scatter plot showing that Ptbp1 knockdown in Neuro2a cells de-represses the inclusion of 24 neuronal exons predicted by motif search in A).
D) RT-PCR analysis showing that Ptbp1 knockdown in Neuro2a cells by three different shRNAs promotes inclusion of Flna exonN.
E) Western blot and quantified signals showing that Ptbp1 knockdown decreases Flna protein level. Data are represented as mean +/− SEM.
F) iCLIP-Seq (re-analysis of (Linares et al., 2015; Masuda et al., 2012)) and RNA-Seq results showing that Ptbp1 binds directly to Flna exonN and its flanking introns in NPCs and C2C12 cells. Red asterisk and dotted line indicate chrX:71,486,253, the homologous nucleotide of human ChrX: 153,594,210 C>T mutation. The arrow indicates direction of transcription.
G) Rbfox1/2/3 expression in Neuro2a cells promotes inclusion of 30 neuronal exons identified by motif analyses in A).
H) RT-PCR results showing that ectopic expression of Rbfox1/2/3 promotes inclusion of Nin exon 29.
I) Overexpression of Rbfox3 in U2OS cells decreases the protein level of centrosomal Nin (green). Data are represented as median +/− SD.
J) Genome browser views of RNA-Seq and HITS-CLIP reads (re-analysis of (Weyn-Vanhentenryck et al., 2014)) showing that Rbfox1/2/3 proteins bind to conserved GCAUG motifs downstream of Nin exon 29 in mouse brains. Vertical red bars on top indicate GCATG sequences.
It has been shown that Ptbp1 binding upstream suppresses exon inclusion (Keppetipola et al., 2012), whereas Rbfox binding downstream enhances exon inclusion (Kuroyanagi, 2009; Wang et al., 2008). We found that Ptbp1 mRNAs and proteins were highly expressed in NPCs (VZ), with Rbfox1/2/3 specifically expressed in neurons (CP, Figures 6B and S6C), consistent with the positional enrichment around neuron-specific exons. Knock down of Ptbp1, but not Ptbp2, de-repressed inclusion of predicted neuronal exons including Flna exonN (Figure 6C–6D, Figure S6D–S6E), and resulted in decreased Flna protein expression (Figure 6E). Moreover, the protein levels of Ptbp1 and Flna were high in E12.5 mouse cortex and dropped over similar time-courses during cortical development (Figure S6F). Analysis of Ptbp1 iCLIP-Seq datasets (Linares et al., 2015; Masuda et al., 2012) revealed that Ptbp1 bound to the alternatively spliced Flna exonN in both NPCs and C2C12 cells (Figure 6F). The mouse homologous nucleotide (chrX:71,486,253) of the PVNH-associated FLNA c.1429+182 G>A mutation lies within a Ptbp1 iCLIP cluster (Figure 6F), suggesting that the human mutation disrupts splicing regulation via PTBP1.
Expression of Rbfox1/2/3 proteins significantly promoted inclusion of predicted neuronal exons (Figure 6G, Figure S6G–S6H), especially Nin exon 29 (Figure 6H), which led to decreased endogenous Ninein at the centrosome (Figure 6I). Re-analysis of Rbfox-1/2/3 HITS-CLIP datasets from P15 mouse brains (Weyn-Vanhentenryck et al., 2014) revealed that Rbfox1 and Rbfox3 bound to a conserved GCAUG motif, and Rbfox2 CLIP tags mapped to a different but adjacent GCAUG motif (Figure 6J and S6I). These results indicate that Rbfox proteins bind to the downstream intron of Nin exon 29 and promote exon inclusion. In summary, Ptbp1 in NPCs suppresses splicing of a family of neuronal exons including Flna exonN, while Rbfox1/2/3 in neurons promotes inclusion of over 120 neuronal exons including Nin exon 29. Thus, Ptbp1 and Rbfox1/2/3 proteins antagonistically regulate neuronal exon inclusion during the NPC-to-neuron transition in vivo.
Rbfox Proteins and Ptbp1 Antagonistically Regulate Neurogenesis in Cerebral Cortical Development
Conditional knockout (cKO) of Ptbp1 in mouse cerebral cortex causes NPCs to detach from the VZ and differentiate prematurely, leading to lethal hydrocephalus, but the pathogenic mechanism and direct downstream targets remain unknown (Shibasaki et al., 2013). We found that Ptbp1 cKO strikingly phenocopied human PVNH caused by FLNA mutations (Figure 7A), and that protein levels of both Flna and its paralog Flnb were significantly decreased in Ptbp1 knockout cells (Figure 7B). Flna exonN was skipped in controls but abnormally included in Ptbp1 cKO (Figure 7C). We identified an unannotated 98 nt exon of Flnb in the homologous location to Flna exonN, and this Flnb exon was also aberrantly included in Ptbp1 cKO (Figure 7C), and is predicted to cause a translational reading frame shift. These results indicate that Ptbp1 represses inclusion of Flna and Flnb poison exons in NPCs, and Ptbp1 loss-of-function leads to decreased Flna/Flnb levels and PVNH.
Figure 7. Ptbp1 and Rbfox Proteins Antagonistically Control Neural Progenitor Cell Differentiation.
A) Immunostaining of E18.5 Ptbp1 conditional knockout (cKO, Nestin-Cre) cerebral cortex showing typical PVNH (white arrow) and thinner ventricular layer of Pax6+ cells (brackets).
B) Western blot of Ptbp1 cKO and control MEF cells showing that proteins level of Ptbp1, Flna and Flnb are decreased in Ptbp1 cKO, while Ptbp2 level is increased.
C) RT-PCR results showing that Ptbp1 cKO in MEF cells de-represses the inclusion of Flna exonN and a 98-nt Flnb exon.
D) –E) Representative images and statistical analysis E) showing that Ptbp1 knockdown (green) at E13.5 results in reduced neural progenitors in the VZ at E15.5. The defect was partially rescued by co-expression of Ptbp1 coding sequence (CDS, red) or FLNA CDS. Numbers in parentheses indicate the number of embryos analyzed. Data are represented as mean +/− SD.
F) –G) Representative images and statistical analysis G) showing that introducing Rbfox3 expression into E13.5 mouse brains resulted in reduced progenitor cells in the VZ and reduced neurons in the CP at E16.5. Ectopic expression of Rbfox3 on top of Ptbp1 knockdown causes a more severe depletion of NPCs in the VZ. Data are represented as mean +/− SD.
H) A working model showing that Rbfox1/2/3 proteins are highly expressed in neurons and promote neuronal exon inclusion (red), while Ptbp1 is expressed in NPCs (blue, VZ) and represses neuronal exon inclusion. Sox2 binds to the promoter region of Ptbp1 (left). Dysregulation of Rbfox1/2/3 mediated AS may lead to brain disorders such as autism and intellectual disability. Mutations that de-represses neuronal exon inclusion in NPCs may result in PVNH (through FLNA).
See also Figure S7.
FLNA mutations cause PVNH in humans (Fox et al., 1998) and Flna knockout mice show microcephaly, defective adherens junctions and reduced cortical progenitors (Feng et al., 2006; Lian et al., 2012). To test whether Flna functions downstream of Ptbp1, we knocked down Ptbp1 in E13.5 mouse cortical progenitors and found this led to fewer NPCs in the VZ due to abnormal cell cycle exit (Figures 7D–7E, and S7A–S7C). Introduction of exogenous FLNA by in utero electroporation (IUE) partially rescued the premature NPC depletion phenotype (Figure 7D–7E), supporting the model that Ptbp1 functions upstream of Flna in cortical neurogenesis.
Ptbp1 represses neuronal fate in tissue culture cells (Xue et al., 2013), and we found that Ptbp1 expression in the developing mouse cortex was highly correlated with Sox2 (Figure S7D). Sox2 is a master transcriptional regulator of NPC identity and indeed, Sox2 bound to the highly active Ptbp1 promoter region in NPCs (main peak = 398nt, fold enrichment= 7.6, p-value =1.0e-16, Figure S7E), suggesting that Ptbp1 is an important target of Sox2 in suppressing neuronal differentiation.
Single knockout of Rbfox1 or Rbfox2 causes minimal structural malformation in mouse cerebral cortex possibly due to gene redundancy (Gehman et al., 2012; Gehman et al., 2011). However, of particular interest is the consequence of Rbfox1/2/3 gain-of-function in NPCs, because Rbfox proteins promote neuronal exon inclusion and have long been used as pan-neuronal markers. Ectopic expression of Rbfox proteins in NPCs not only significantly decreased the number of VZ NPCs but also dramatically reduced the number of neurons in the CP (Figure 7F–7G). Furthermore, combining Rbfox3 expression with Ptbp1 knock down led to a more severe depletion of VZ NPCs (Figure 7F–7G). These data indicate that Rbfox1/2/3 and Ptbp1 antagonistically regulate neurogenesis in the developing cerebral cortex (Figure 7H).
DISCUSSION
Here we show that widespread alternative exon usage during the NPC-to-neuron transition in vivo is critical for mouse and human brain development. Neuronal fate and neuronal exon inclusion are antagonistically regulated by Ptbp1 and Rbfox1/2/3, which are expressed in NPCs and neurons, respectively. While Rbfox1/2/3-induced splicing causes Nin to be translocated from the centrosome in NPCs to non-centrosomal loci in neurons, the expression of Ptbp1 is required for apical progenitor lamination through maintaining Flna and Flnb expression in NPCs.
Perhaps most remarkable in our study is the fact that two opposite and extreme alternative splicing events in a single target gene, Ninein, appear sufficient to differentiate NPCs to neurons by removing CEP170- and CEP250-binding domains/exons. This alternative splicing mechanism of Nin may also explain previously described microtubule re-organization phenomena during epidermal differentiation (Lechler and Fuchs, 2007) and could potentially regulate epidermal differentiation itself. While our unbiased motif analysis revealed the robust enrichment of Ptbp1/2- and Rbfox1/2/3-binding sequences flanking neuronal exons during the NPC-to-neuron transition, other RBPs may synergistically regulate neuronal exon usage in a context-dependent manner. ELAVL, NOVA, nSR100 and STAR proteins have been reported to regulate specific AS events during cerebral cortical development (Calarco et al., 2009; Darnell, 2013; Iijima et al., 2011), and some of these factors are also differentially expressed between cortical NPCs and neurons.
To our surprise, the brain-specific Ptbp1 knockout is a mouse model for PVNH (Figure 7A and (Shibasaki et al., 2013)). While FLNA mutations were linked to human PVNH two decades ago, Flna knockout mice only recapitulate the cardiovascular defects in human patients but do not exhibit PVNH (Feng et al., 2006), likely due to redundant functions of Flnb. Ptbp1 loss disrupts both Flna and Flnb, causing radial glia to detach from apical surface, and forming PVNH. These results demonstrate the essential roles of the Ptbp1-Filamin axis in maintaining NPCs and neuroepithelial structure.
As expected from the cell type-specific splicing of FLNA, the intronic FLNA-as mutation de-represses exonN inclusion and leads to a unique presentation of PVNH with the absence of non-CNS manifestations, suggesting the preferential use of this alternative-splicing mechanism in brain. The homologous nucleotide of the FLNA c.1429+182 G>A mutation lies within a Ptbp1 iCLIP cluster (Figure 6F), suggesting that the human mutation disrupts splicing regulation via PTBP1. In agreement with this, a computational model of Ptbp1 binding based on iCLIP clusters reported that interspersed G residues are well tolerated, but that A residues always impair Ptbp1 binding (Han et al., 2014).
This study reveals that cell type-specific alternative splicing—frequently associated with alternative stop codons and NMD—is widespread in the central nervous system at early developmental stages, and provides functional insights into developmental roles for some of these alternative exons and their upstream splicing regulators. The active usage of many other cell type-specific poison exons demonstrates not only the demand for larger efforts in genetic testing of their flanking non-coding elements, but also the potential of uncovering more master splicing regulators that may play important roles in neurological disorders.
METHODS AND RESOURCES
CONTACT FOR REAGENT AND RESOURCE SHARING
Please contact C.A.W. ([email protected]) for reagents and resources generated in this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse Maintenance
Mouse protocols were reviewed and approved by Institutional Animal Care and Use Committee (IACUC) at Boston Children’s Hospital (BCH) and all colonies were maintained following animal research guidelines at BCH. The Tbr2-EGFP transgenic mouse line, for which an EGFP-PolyA cassette was inserted in front of the translational start codon in a FVB/NTac mouse stain, was obtained from GENSAT (Gong et al., 2003) and crossed to the FVB inbreed strain for at least two generations. Heterozygous transgenic mice did not display any obvious developmental defects. Dorsal cerebral cortices were dissected from embryonic day 14.5 (E14.5) heterozygous Tbr2-EGFP transgenic embryos and pooled for splicing analyses without distinguishing genders.
Human Subjects
All human protocols were reviewed and approved by the institutional review board of BCH. Informed consent was obtained from all subjects involved in this study or from parents of those who were younger than 18 years old. PVNH cases screened in this study were recruited from diverse ethic groups and a Caucasian pedigree carrying the FLNA-as mutation is described. Mutations were detected with DNA extracted from blood samples. Splicing analyses of affected individuals were performed with RNA samples extracted from immortalized lymphoblasts.
Isolation of Primary Cells from the Developing Mouse Cerebral Cortex
Dissected brain tissues are dissociated with the Papain Dissociation System (Worthington). Single cell suspension (Neural Basal medium supplemented with 2% B27, 1% N2, 1% Penicillin Streptomycin (PenStrep), 10ng/ml EGF, 10ng/ml FGF2, 0.5% FBS and 0.25% HEPES) was cooled on ice and immediately sorted on the FACSAria II (BD). For immunostaining, sorted cells were plated in Lab-Tek chamber slides (Thermo) and cultured for 4 hours before fixation.
Tissue Culture Cells
Cell lines including Neuro2a (a neuroblastoma cell line with a NPC-like state), Hela and Ptbp1 cKO MEF cells were used for transfection (Lipofectamine 2000, Life Tech) or lentiviral transduction. Lentiviral vectors were packaged and infected following the The RNAi Consortium (TRC) protocols. Briefly, the envelop plasmid (pMD2.G, Addgene, 12259), packaging plasmid (psPAX2, Addgene, 12260), and pLKO.1 based vectors were cotransfected into HEK 293T/17 (ATCC, CRL-11268), and viral particles were harvested 42 hours post-trasfection. Ptbp1 cKO MEF cells were transduced with lentiviral particles carrying control plasmid or CMV-Cre for protein and RNA analyses. To generate NINEIN knockout cell line, pX459 (Addgene 48139) carrying guide sequences were transfected into Hela cells and puromycin (5µg/ml) selected colonies were screened for mutant alleles.
METHOD DETAILS
RNA Sequencing analysis
Total RNA was extracted using RNeazy Mini Kit (Qiagen) and treated with Turbo DNase (Ambion). RINs measured by Bioanalyzer (Agilent) for all samples were above 9.0. 500–2000ng total RNA was treated with Ribo-Zero Gold (Epicentre), and mRNA was enriched using Oligo(dT)25 Dynabeads (Life Tech). RNA-Seq libraries (2 replicates each for VZ and non-VZ cells, n or number of embryos= 7 and 11 for VZ, n=7 and 8 for non-VZ replicates) were prepared using ligation-based directional RNA-Seq kit (Bioo Scientific), and sequenced on Illumina HiSeq2000. 36 to 42 million single reads (50bp) were obtained for each replicate, trimmed, aligned (mm9, Tophat-2.0.7, allowing 2 mismatches) and analyzed using the Tuxedo protocol (Kim et al., 2013; Trapnell et al., 2012). Gene expression levels are presented as FPKM (Fragments Per Kilobase of exon per Million reads). Gene ontology analyses were performed using the DAVID online tools (Huang da et al., 2009).
Alternative Splicing Analysis
Aligned bam files (mm9) were analyzed using MISO (version 0.4.6). PSI values of each non-VZ replicate were compared to each VZ replicate, only alternative splicing events that were consistent across all comparisons were considered for downstream analysis. Additional RNA-Seq datasets of laser microdissected cortical tissues (Ayoub et al., 2011; Fietz et al., 2012) were analyzed using the same pipeline. Sashimi plots of RNA-Seq reads were generated in Integrative Genomics Viewer (IGV) (Robinson et al., 2011; Thorvaldsdottir et al., 2013). To study alternative splicing changes during human brain development, published RNA-Seq datasets (Fietz et al., 2012) on laser microdissected fetal human cortical tissues across GW13–16 were aligned to human genome (hg19) and analyzed using the MISO pipeline. Alternatively spliced human exons (|ΔPSI|≥10%, and Bayes factor ≥5) were lifted over to the mouse genome (mm9) and compared to splicing events identified in E14.5 mouse cortex (Figure 1D). RNA-Seq datasets from the ENCODE project were analyzed for tissue distribution of alternative exons.
Motif Analysis
We performed DREME (Bailey, 2011) motif discovery in each of the four regions around differentially spliced cassette exons in mouse: the first 200 nts (i.e. 5’ end) of the upstream intron, the last 200 nts (i.e. 3’ end) of the upstream intron, the first 200 nts of the downstream intron, and the last 200 nts of the downstream intron. For each region, we used exons with higher PSI in neurons as foreground, and exons with higher PSI in NPCs as background, and verse versa. DREME (version 4.9.0) were then run with the option ‘-norc –e 0.05’ to identify significant motifs in each region.
Chip-Seq Analysis
Chip-Seq reads for Sox2 (Lodato et al., 2013) and H3K27ac (Creyghton et al., 2010) on NPCs cultured in vitro were trimmed, and aligned to the mouse genome (mm9) with Bowtie 2.2.4 (Langmead and Salzberg, 2012) allowing zero or one mismatch. Mapped reads were visualized in IGV. Peaks were called using MACS 2.1.0 (Zhang et al., 2008) and statistical results were presented.
Molecular Cloning
pCAGIG (Addgene, 11159) vector and its derivatives were used for ectopic expression of genes in the neuronal system. First, the multiple cloning sites (MCS) between EcoR I and Not I was replaced with EcoR I-ATG(start codon)-HA(1x)-Flag(1x)-Asc I-Not I. Next, the EGFP expression cassette between Msc I and Fse I was replaced with either an mCherry or Puromycin resistance cassette. Coding or genomic sequences of specific genes (e.g., genomic sequences of WT human FLNA between exon 8 and exon 12 and the same sequence carrying the c.1429+182 G>A mutation) were PCR amplified and inserted between Asc I and Not I sites. To knockdown Ptbp1 and Ptbp2 in Neuro2a cells, oligos were annealed and cloned into pLKO.1 TRC cloning vector (Addgene, 10878) between EcoR I and Age I sites. To knockdown Ptbp1 expression in mouse brains, effective hairpins tested in Neuro2a cells (#3) and a control hairpin were end-modified and cloned into the pLL3.7 vector (Addgene, 11795) between Hpa I and Xho I sites. For reverse transcription and polymerase chain reaction (RT-PCR), cDNAs from independent samples were amplified with Phusion (NEB) for 26–30 cycles and analyzed on agarose gels or Novex TBE gels (Life Tech). qRT-PCR (quantitative RT-PCR) were carried out using SYBR Green (ABI). Oligo sequences are listed in Table S5, and plasmids are listed in Table S6.
Protein Analysis
Protein domains were analyzed using Pfam (Finn et al., 2014). 3-D protein structures were predicted on the SWISS-MODEL and Phyre2 servers (Kelley and Sternberg, 2009). Protein lysates were resolved on SDS-PAGE gels and Western Blots were carried out using the Licor Odyssey system. For immunofluorescence staining (IF), embryonic mouse brains were dissected out, fixed in 4% paraformaldehyde, and coronal sections were obtained using a Vibratome or Cryostat (Leica Biosystems). Tissue sections or fixed tissue culture cells were rinsed twice in 1x PBS with 0.2% Triton X-100 (PBST) and incubated at room temperature in a blocking solution (PBST and 4% normal donkey serum), followed by incubation with primary antibodies at 4°C overnight. Samples were then washed 3 times with PBST and incubated with fluorescence conjugated secondary Alexa antibodies (Life Technologies) at room temperature for 2 hours. Slides were mounted with Fluoromount G (Southern Biotech) and imaged on Imager M2 (Carl Zeiss). Primary antibodies are listed in Key Resource Table.
In utero Electroporation (IUE)
IUE was performed following standard protocols (Saito, 2006). In brief, a timed pregnant CD-1 mouse (E13.5) was anesthetized with isoflurane, laid on a 37°C warm plate, the uterine horns exposed and bathed in warm 1x HBSS (1% Pen/Strep), and ~1 µl of plasmid DNA (1–2µg/ µl) mixed with Fast Green was microinjected into the lateral ventricles of embryos using a glass micropipette (Drummond Scientific). Five 50 ms pulses of 30–50 mV at 950-ms intervals were delivered across the uterus with a pair of electrode paddles placed on each side of the embryo’s head using a square-pulse electroporator (BTX, ECM 830). Following electroporation, the uterus was repositioned back into the abdominal cavity and the wound was surgically sutured. The whole process was performed in a sterile environment. After surgery, the animals were closely monitored until they recovered and resumed normal activity. Pair-cell assays were performed following published protocols (Shen et al., 2002). Briefly, in utero electroporated NPCs expressing ectopic Nin isoforms were dissociated, FACS sorted, plated at clonal density and their progeny pair classified as progenitors (Sox2+; Tuj1−), neurons (Sox2−; Tuj1+) or a mixture.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses for differential gene expression and splicing changes (Figures 1, 2 and 6) were performed in R and MISO. Levels of significance were calculated with Student’s t-test for Figure 6I, and with Hypergeometric Test for Figure S6B. Isoform abundance on DNA gels was measured in Photoshop and normalized with DNA length. For statistical analyses of cell fates (Figure 4 and Figure 7), five to thirteen positive embryos were analyzed for each condition (specific numbers of analyzed embryos are indicated in each figure), and three to four sections were examined for each embryo. Percentages presented Figure 4 and Figure 7 were calculated by dividing the number of EGFP+ cells in each zone (eg, VZ, IZ/SVZ and CP) by the total number of EGFP+ cells of all zones on the same images. Percentages of cells in the VZ were compared using one-way ANOVA in GraphPad Prism, and means +/− (standard deviation, SD) are presented. In all figures: *, p-value < 0.05; **, p-value < 0.01; ***, p-value < 0.001.
Supplementary Material
A) Immunostaining of E10.5, E12.5 and E14.5 Tbr2-EGFP mouse dorsal cerebral cortex with anti-EGFP (green), anti-Tbr2 (red) and anti-Sox2 (magenta). White arrows indicate that (EGFP+) cells in the VZ/SVZ are (Tbr2+; Sox2−). Scale bar, 20 µm.
B) Quantitative RT-PCR results showing that Sox2 and Pax6 mRNAs are enriched in FACS sorted VZ cells.
C) RT-PCR results showing that the neuronal exon 4 of REST is enriched in FACS sorted non-VZ cells and depleted from sorted VZ cells.
D) Immunostaining of FACS sorted VZ and non-VZ cells with anti-Sox2, anti-Tbr2 and anti-DCX. Scale bar, 100 µm.
E) Statistic analysis of D) showing that sorted VZ cells are mostly Sox2 (+), and that sorted non-VZ cells are Dcx postive.
F) Gene ontology analysis of differentially expressed genes (over 4 fold difference, p < 0.01) showing that cell cycle related genes are overrepresented in sorted VZ cells, and synaptic and neuron differentiation genes are enriched in sorted non-VZ cells.
G) A Venn diagram showing the number of shared alternative splicing events that we identified in three independent RNA-Seq datasets.
H) A cumulative dot plot showing the size distribution of mouse SEs.
I) Heatmap and unsupervised clustering of PSI values for alternative exons (each row or line) between human VZ, iSVZ, oSVZ and CP (magenta represent high PSI value and light blue showing low PSI values).
J) Pie chart showing alternative exons that are specifically included or skipped in each of VZ, iSVZ, oSVZ and CP samples.
K) Histograms showing the size of human SEs (x axis) and the percentages of SEs that cause an in-frame insertion or a frame shift (y axis).
A) Gene ontology analysis of human genes that were differentially spliced between GW13-16 fetal human NPCs and neurons, showing that genes related to cytoskeleton and other biological functions were overrepresented.
B) A Venn diagram showing that alternative exons in 38 of 66 (58%) cytoskeleton related mouse SEs are also alternatively spliced in the developing human cerebral cortex.
C) KEGG pathway analysis of alternatively spliced genes revealed their functions (red stars) in regulating tight junctions (p-value=0.002).
D) –E) Additional RT-PCR analyses of alternatively spliced exons in microtubule- (C) and actin- (D) related genes in mice.
A) Number of amino acids affected by alternative splicing and their impact on protein domains, with bars showing the median size. Note that short SEs including microexons (5 AA or shorter) tend to insert extra AA into protein domains.
B) Effects of alternative splicing on protein domains. Gene names are followed by ?PSI values. Differential sequences are colored light red (neuron specific) or light blue (NPC specific). Differential protein domains are shaded red (neuron) or cyan (NPC).
C) Representative neuron specific cassette exons inserting extra amino acids (red) into defined proteins domains (cyan). Left to right: 32 amino acids insert to the spectrin-binding ZU5 domain of Ank2; 12 amino acids insert to the ELMO inhibitory domain (EID) of Elmo2; 8 amino acids insert into the 15th immunoglobin like repeat of Flna; 6 amino acids insert into the growth-arrest-specific protein 2 (GAS2) domain of Macf1.
D) Prolonged and high expression of Nin-Neuron isoform diffuses from centrosomal area to cytoplasm.
E) Endogenous non-centrosomal Nin associates with microtubules in Caco2 cells.
F) Endogenous Nin localizes to non-centrosomal microtubules in primary mouse hippocampal neurons.
G) Western blot results showing that siRNAs against CEP250 efficiently knocked down CEP250 levels.
H) EGFP-Nin-NPC, but EGFP-Nin-Neuron, co-localizes with CEP170.
A) FPKM values showing that NIN expression is higher in CP than in VZ, iSVZ and oSVZ.
B) PSI values showing that NIN exon18 (2139 nt) is specifically included in VZ and the neuronal exon29 (61 nt) is specifically included in neurons.
C) Introducing the Nin-Neuron isoform, but not the Nin-NPC isoform, into the E13.5 mouse brain leads to fewer NPCs in the VZ and fewer neurons in the CP at E16.5.
D) The guide sequence used to generate NIN knockout cells and the p.Arg107Thrfs*16 early truncation mutation.
A) Alignment of mouse Flna and human FLNA exonNs (red) and their upstream and downstream exons, showing that the alternative exonNs and the embedded premature stop codons (dashed red line box) are conserved.
B) Multiple alternatively spliced transcripts around FLNA exonN.
A) A Venn diagram showing numbers of alternative exons that have upstream Ptbp- and/or downstream Rbfox-binding sites.
B) Venn diagrams showing Rbfox and Ptbp1 iCLIP peaks associated with predicted targets in A) using previously published iCLIP/HITS-CLIP datasets (Linares et al., 2015; Weyn-Vanhentenryck et al., 2014). .
C) Immuno-fluorescence staining (green) showing that Rbfox1 is highly expressed in the CP (left), and Ptbp1 (magenta) is specifically expressed in the VZ (right).
D) Western blot showing that different shRNAs against mouse Ptbp1 (3) or Ptbp2 (2) efficiently knock down target protein expression in Neuro2a cells.
E) RT-PCR analyses showing that Ptbp1 knockdown promoted the inclusion of neuronal exons, with PSI values shown below.
F) Western-blot of cerebral cortex lysates showing that the protein levels of Flna and Ptbp1 peaks during early cortex development (E12.5) and drops at later stages.
G) Western Blot showing ectopic expression of Rbfox1/2/3 isoforms in Neuro2a cells.
H) RT-PCR analyses show that ectopic expression of Rbfox1/2/3 increases neuronal exon inclusion.
I) HITS-CLIP (re-analysis of datasets reported by an independent study (Masuda et al., 2012) ) and RNA-seq results showing that Rbfox1/2/3 proteins bind to conserved GCAUG sequences downstream of Nin exon 29.
A) Ptbp1 shRNA knockdown (anti-EGFP) depleted neural progenitors from VZ.
B) –C) Representative images and statistical analysis (D) showing that Ptbp1 knockdown (anti-EGFP, red) promotes neural progenitors to exit cell cycle (BrdU+; Ki67−). y axis represents the proportion of (EGFP+; BrdU+; Ki67−) cells over (EGFP+; BrdU+) cells. Data are represented as mean ± SEM.
D) Ptbp1 co-localizes with Sox2 but not Tbr2 in E14.5 mouse cerebral cortex.
E) Sox2 and H3K27ac Chip-Seq reads showing that Sox2 binds to the active Ptbp1 promoter in neural progenitor cells.
Acknowledgments
We thank B. Barry and J. Partlow for clinical information, Y.J. Yang, A. Lam, D. Gonzalez and A. Rozzo for technical assistance, M. Bornens, V. Sheen and G. Lian for reagents, the Orchestra research computing team at Harvard for computing resources, N. Francis of the IDDRC Flow Cytometry Core for technical assistance, B. Bae, X. Cai, G. Evrony, R. S. Hill, E. Lim, M. Lodato, M. Johnson and Walsh lab members for helpful discussions. We thank Qiufu Ma and Xi He for critical comments on the manuscript. This work was supported by the Manton Center for Orphan Disease Research and grants from the NINDS (R01-NS035129 and R01-NS032457) to C.A.W., NIMH grant (K01-MH109747) and Charles A. King Trust Fellowship to X.Z, HHMI International Student Research Fellowship to X.W., NSF graduate fellowship (DGE1144152) to J.F., and NIGMS (R01-GM34277) and NCI (P01-CA42063) to P.A.S. C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
DATA AND SOFTWARE AVAILABILITY
Data Resources
The accession number for the RNA sequencing data reported in this paper is NCBI Gene Expression Omnibus: GSE76198.
AUTHOR CONTRIBUTIONS
X.Z. conceived and developed the project, and performed most wet-lab and bioinformatics experiments. X.W. assisted in MISO analyses and performed RNA motif search. M.H.C. recruited PVNH cases. A.K. and J.F.R. performed Ninein co-IP and related IF. J.F and P.V.K analyzed single cell RNA-seq data with advice from X.Z. R.D. assisted in screening FLNA mutations. M.O. and N.Y. provided Ptbp1 cKO samples. J.M. tested reagents. D.L.B. and P.A.S provided guidance on splicing and iCLIP analyses. X.Z. and C.A.W designed the study and wrote the paper with input from all authors.
REFERENCES
- Ayoub AE, Oh S, Xie Y, Leng J, Cotney J, Dominguez MH, Noonan JP, Rakic P. Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14950–14955. doi: 10.1073/pnas.1112213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird DH, Myers KA, Mogensen M, Moss D, Baas PW. Distribution of the microtubule-related protein ninein in developing neurons. Neuropharmacology. 2004;47:677–683. doi: 10.1016/j.neuropharm.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, Garcia-Moreno F, Molnar Z, Margulies EH, Ponting CP. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla K, Phillips HA, Crawford J, McKenzie OL, Mulley JC, Eyre H, Gardner AE, Kremmidiotis G, Callen DF. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O’Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. RNA protein interaction in neurons. Annual review of neuroscience. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber A, Lafranchi SH, Maliga Z, Lui JC, Moon JE, McDeed C, Henke K, Zonana J, Kingman GA, Pers TH, et al. Novel microcephalic primordial dwarfism disorder associated with variants in the centrosomal protein ninein. The Journal of clinical endocrinology and metabolism. 2012;97:E2140–E2151. doi: 10.1210/jc.2012-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci. 2005;118:1565–1575. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- Dillman AA, Hauser DN, Gibbs JR, Nalls MA, McCoy MK, Rudenko IN, Galter D, Cookson MR. mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nature neuroscience. 2013;16:499–506. doi: 10.1038/nn.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Salathia N, Liu R, Kaeser GE, Yung YC, Herman JL, Kaper F, Fan JB, Zhang K, Chun J, et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nature methods. 2016;13:241–244. doi: 10.1038/nmeth.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, Kwiatkowski DJ, Walsh CA. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19836–19841. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA, Lachmann R, Brandl H, Kircher M, Samusik N, Schroder R, Lakshmanaperumal N, Henry I, Vogt J, Riehn A, et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11836–11841. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic acids research. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, Scheffer IE, Dobyns WB, Hirsch BA, Radtke RA, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, et al. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Meera P, Stoilov P, Shiue L, O’Brien JE, Meisler MH, Ares M, Jr, Otis TS, Black DL. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes & development. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Jr, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nature genetics. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. The Journal of cell biology. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nature reviews Neuroscience. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Stoilov P, Linares AJ, Zhou Y, Fu XD, Black DL. De novo prediction of PTBP1 binding and splicing targets reveals unexpected features of its RNA recognition and function. PLoS computational biology. 2014;10:e1003442. doi: 10.1371/journal.pcbi.1003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huttner WB, Kelava I, Lewitus E. Progenitor networking in the fetal primate neocortex. Neuron. 2013;80:259–262. doi: 10.1016/j.neuron.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Iijima T, Wu K, Witte H, Hanno-Iijima Y, Glatter T, Richard S, Scheiffele P. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147:1601–1614. doi: 10.1016/j.cell.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallieres M, Tapial J, Raj B, O’Hanlon D, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, Walsh CA. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nature neuroscience. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nature methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. The Journal of cell biology. 2007;176:147–154. doi: 10.1083/jcb.200609109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nature reviews Neuroscience. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Li YI, Sanchez-Pulido L, Haerty W, Ponting CP. RBFOX and PTBP1 proteins regulate the alternative splicing of micro-exons in human brain transcripts. Genome research. 2015;25:1–13. doi: 10.1101/gr.181990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian G, Lu J, Hu J, Zhang J, Cross SH, Ferland RJ, Sheen VL. Filamin a regulates neural progenitor proliferation and cortical size through Wee1-dependent Cdk1 phosphorylation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:7672–7684. doi: 10.1523/JNEUROSCI.0894-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares AJ, Lin CH, Damianov A, Adams KL, Novitch BG, Black DL. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. eLife. 2015;4:e09268. doi: 10.7554/eLife.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato MA, Ng CW, Wamstad JA, Cheng AW, Thai KK, Fraenkel E, Jaenisch R, Boyer LA. SOX2 co-occupies distal enhancer elements with distinct POU factors in ESCs and NPCs to specify cell state. PLoS genetics. 2013;9:e1003288. doi: 10.1371/journal.pgen.1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda A, Andersen HS, Doktor TK, Okamoto T, Ito M, Andresen BS, Ohno K. CUGBP1 and MBNL1 preferentially bind to 3’ UTRs and facilitate mRNA decay. Sci Rep. 2012;2:209. doi: 10.1038/srep00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama Y, Hayashi K. Relocalization of a microtubule-anchoring protein, ninein, from the centrosome to dendrites during differentiation of mouse neurons. Histochem Cell Biol. 2009;132:515–524. doi: 10.1007/s00418-009-0631-z. [DOI] [PubMed] [Google Scholar]
- Raj B, Blencowe BJ. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron. 2015;87:14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Raj B, O’Hanlon D, Vessey JP, Pan Q, Ray D, Buckley NJ, Miller FD, Blencowe BJ. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Molecular cell. 2011;43:843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nature protocols. 2006;1:1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- Shibasaki T, Tokunaga A, Sakamoto R, Sagara H, Noguchi S, Sasaoka T, Yoshida N. PTB deficiency causes the loss of adherens junctions in the dorsal telencephalon and leads to lethal hydrocephalus. Cereb Cortex. 2013;23:1824–1835. doi: 10.1093/cercor/bhs161. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nature reviews Neuroscience. 2016;17:265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell reports. 2014;6:1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Weyn-Vanhentenryck SM, Wu J, Sloan SA, Zhang Y, Chen K, Wu JQ, Barres BA, Zhang C. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3445–3450. doi: 10.1073/pnas.1502849112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Immunostaining of E10.5, E12.5 and E14.5 Tbr2-EGFP mouse dorsal cerebral cortex with anti-EGFP (green), anti-Tbr2 (red) and anti-Sox2 (magenta). White arrows indicate that (EGFP+) cells in the VZ/SVZ are (Tbr2+; Sox2−). Scale bar, 20 µm.
B) Quantitative RT-PCR results showing that Sox2 and Pax6 mRNAs are enriched in FACS sorted VZ cells.
C) RT-PCR results showing that the neuronal exon 4 of REST is enriched in FACS sorted non-VZ cells and depleted from sorted VZ cells.
D) Immunostaining of FACS sorted VZ and non-VZ cells with anti-Sox2, anti-Tbr2 and anti-DCX. Scale bar, 100 µm.
E) Statistic analysis of D) showing that sorted VZ cells are mostly Sox2 (+), and that sorted non-VZ cells are Dcx postive.
F) Gene ontology analysis of differentially expressed genes (over 4 fold difference, p < 0.01) showing that cell cycle related genes are overrepresented in sorted VZ cells, and synaptic and neuron differentiation genes are enriched in sorted non-VZ cells.
G) A Venn diagram showing the number of shared alternative splicing events that we identified in three independent RNA-Seq datasets.
H) A cumulative dot plot showing the size distribution of mouse SEs.
I) Heatmap and unsupervised clustering of PSI values for alternative exons (each row or line) between human VZ, iSVZ, oSVZ and CP (magenta represent high PSI value and light blue showing low PSI values).
J) Pie chart showing alternative exons that are specifically included or skipped in each of VZ, iSVZ, oSVZ and CP samples.
K) Histograms showing the size of human SEs (x axis) and the percentages of SEs that cause an in-frame insertion or a frame shift (y axis).
A) Gene ontology analysis of human genes that were differentially spliced between GW13-16 fetal human NPCs and neurons, showing that genes related to cytoskeleton and other biological functions were overrepresented.
B) A Venn diagram showing that alternative exons in 38 of 66 (58%) cytoskeleton related mouse SEs are also alternatively spliced in the developing human cerebral cortex.
C) KEGG pathway analysis of alternatively spliced genes revealed their functions (red stars) in regulating tight junctions (p-value=0.002).
D) –E) Additional RT-PCR analyses of alternatively spliced exons in microtubule- (C) and actin- (D) related genes in mice.
A) Number of amino acids affected by alternative splicing and their impact on protein domains, with bars showing the median size. Note that short SEs including microexons (5 AA or shorter) tend to insert extra AA into protein domains.
B) Effects of alternative splicing on protein domains. Gene names are followed by ?PSI values. Differential sequences are colored light red (neuron specific) or light blue (NPC specific). Differential protein domains are shaded red (neuron) or cyan (NPC).
C) Representative neuron specific cassette exons inserting extra amino acids (red) into defined proteins domains (cyan). Left to right: 32 amino acids insert to the spectrin-binding ZU5 domain of Ank2; 12 amino acids insert to the ELMO inhibitory domain (EID) of Elmo2; 8 amino acids insert into the 15th immunoglobin like repeat of Flna; 6 amino acids insert into the growth-arrest-specific protein 2 (GAS2) domain of Macf1.
D) Prolonged and high expression of Nin-Neuron isoform diffuses from centrosomal area to cytoplasm.
E) Endogenous non-centrosomal Nin associates with microtubules in Caco2 cells.
F) Endogenous Nin localizes to non-centrosomal microtubules in primary mouse hippocampal neurons.
G) Western blot results showing that siRNAs against CEP250 efficiently knocked down CEP250 levels.
H) EGFP-Nin-NPC, but EGFP-Nin-Neuron, co-localizes with CEP170.
A) FPKM values showing that NIN expression is higher in CP than in VZ, iSVZ and oSVZ.
B) PSI values showing that NIN exon18 (2139 nt) is specifically included in VZ and the neuronal exon29 (61 nt) is specifically included in neurons.
C) Introducing the Nin-Neuron isoform, but not the Nin-NPC isoform, into the E13.5 mouse brain leads to fewer NPCs in the VZ and fewer neurons in the CP at E16.5.
D) The guide sequence used to generate NIN knockout cells and the p.Arg107Thrfs*16 early truncation mutation.
A) Alignment of mouse Flna and human FLNA exonNs (red) and their upstream and downstream exons, showing that the alternative exonNs and the embedded premature stop codons (dashed red line box) are conserved.
B) Multiple alternatively spliced transcripts around FLNA exonN.
A) A Venn diagram showing numbers of alternative exons that have upstream Ptbp- and/or downstream Rbfox-binding sites.
B) Venn diagrams showing Rbfox and Ptbp1 iCLIP peaks associated with predicted targets in A) using previously published iCLIP/HITS-CLIP datasets (Linares et al., 2015; Weyn-Vanhentenryck et al., 2014). .
C) Immuno-fluorescence staining (green) showing that Rbfox1 is highly expressed in the CP (left), and Ptbp1 (magenta) is specifically expressed in the VZ (right).
D) Western blot showing that different shRNAs against mouse Ptbp1 (3) or Ptbp2 (2) efficiently knock down target protein expression in Neuro2a cells.
E) RT-PCR analyses showing that Ptbp1 knockdown promoted the inclusion of neuronal exons, with PSI values shown below.
F) Western-blot of cerebral cortex lysates showing that the protein levels of Flna and Ptbp1 peaks during early cortex development (E12.5) and drops at later stages.
G) Western Blot showing ectopic expression of Rbfox1/2/3 isoforms in Neuro2a cells.
H) RT-PCR analyses show that ectopic expression of Rbfox1/2/3 increases neuronal exon inclusion.
I) HITS-CLIP (re-analysis of datasets reported by an independent study (Masuda et al., 2012) ) and RNA-seq results showing that Rbfox1/2/3 proteins bind to conserved GCAUG sequences downstream of Nin exon 29.
A) Ptbp1 shRNA knockdown (anti-EGFP) depleted neural progenitors from VZ.
B) –C) Representative images and statistical analysis (D) showing that Ptbp1 knockdown (anti-EGFP, red) promotes neural progenitors to exit cell cycle (BrdU+; Ki67−). y axis represents the proportion of (EGFP+; BrdU+; Ki67−) cells over (EGFP+; BrdU+) cells. Data are represented as mean ± SEM.
D) Ptbp1 co-localizes with Sox2 but not Tbr2 in E14.5 mouse cerebral cortex.
E) Sox2 and H3K27ac Chip-Seq reads showing that Sox2 binds to the active Ptbp1 promoter in neural progenitor cells.