Abstract
Development of biofluid-based molecular diagnostic tests for cancer is an important step towards tumor characterization and real-time monitoring in a minimally invasive fashion. Extracellular vesicles (EVs) are released from tumor cells into body fluids and can provide a powerful platform for tumor biomarkers because they carry tumor proteins and nucleic acids. Detecting rare point mutations in the background of wild-type sequences in biofluids such as blood and cerebrospinal fluid (CSF) remains a major challenge. Techniques such as BEAMing (beads, emulsion, amplification, magnetics) PCR and droplet digital PCR (ddPCR) are substantially more sensitive than many other assays for mutant sequence detection. Here, we describe a novel approach that combines biofluid EV RNA and BEAMing RT-PCR (EV-BEAMing), as well droplet digital PCR to interrogate mutations from glioma tumors. EVs from CSF of patients with glioma were shown to contain mutant IDH1 transcripts, and we were able to reliably detect and quantify mutant and wild-type IDH1 RNA transcripts in CSF of patients with gliomas. EV-BEAMing and EV-ddPCR represent a valuable new strategy for cancer diagnostics, which can be applied to a variety of biofluids and neoplasms.
Keywords: BEAMing PCR, biomarkers, cancer, extracellular vesicles, droplet digital PCR
Introduction
There has been growing interest in cancer diagnostics using circulating tumor-derived nucleic acids as a source for tumor biomarkers. This approach is particularly attractive because it allows detection of genetic mutations in the tumor as well as quantification of mutant sequences present in the sample. Biofluid-based analysis is less invasive than performing tumor biopsy, extends the range of a single biopsy by offering the potential to mirror the genetic diversity within a tumor, and also enables longitudinal measurements to monitor genetic changes in tumor over time.1,2
A new promising source of biofluid-based biomarkers is extracellular vesicles (EVs); they are actively released from normal as well as cancer cells both in vitro and in vivo,3 with upregulated release from many tumor cells, as compared with normal cells.3,4,5,6 They include several types of membrane-bound vesicles, such as exosomes, shed EVs, oncosomes and apoptotic blebs.7 EVs contain a diverse repertoire of genetic material and protect their cargo from the destructive action of enzymes, such as RNases,4 thus enabling a stable enrichment of nucleic acids from biofluids. RNA from glioblastoma (GBM) serum EVs has previously been used to detect the EGFRvIII mutation.4 Tumor-derived EVs are also found in the cerebrospinal fluid (CSF),8 and may provide valuable biomarker information about the tumor origins and status. Several diagnostic techniques utilize CSF for analysis; however, most of them do not provide any molecular or genetic information on malignancies. In vivo techniques, including CSF cytology, clinical findings and neuro-imaging, as well as in vitro techniques like immunocytochemistry, flow cytometry and PCR have so far only been informative for leptomeningeal cancers, in which tumor cells have entered and disseminated within CSF.9
Highly sensitive technologies must be used when detecting mutations in the background of wild-type sequence and the optimal assay may vary according to the type of genetic alteration to be detected. For example, large gene/transcript deletions like EGFRvIII are relatively easy to detect in the background of wild-type sequences using well optimized allele-specific PCR assays.4 In contrast, single nucleotide mutations are very difficult to pick up when the fraction of mutated transcripts is low in a background of wild-type sequences, and even well optimized allele-specific PCRs can rarely detect mutations at a frequency of less than 1%.10 To increase the sensitivity of EV RNAs as tumor biomarkers, we adapted a well established and highly sensitive form of digital PCR known as BEAMing (Beads, Emulsions, Amplification, Magnetics)11 to interrogate the mRNA within EVs for the presence of tumor specific mutations. BEAMing PCR is a highly sensitive and quantitative technique that has been successfully applied to the mutational analysis of circulating DNA even when the mutation is as rare as 0.01%.12,13,14 We also used a droplet digital PCR (ddPCR) technology, which creates millions of uniform 5 picoliter-volume droplets per sample for counting applications including rare allele detection and validation in tumor and cell-free plasma,15,16,17 and assessment of DNA quality before sequencing.18 Neither of these technologies have previously been used to quantify rare mutations at the RNA level or been tested on EV nucleic acids.
The IDH1 mutation was chosen for analysis because of its high relevance in tumor pathogenesis. Analysis was carried out for wild-type (G395) and mutant (A395) sequences in the mRNA transcripts for isocitrate dehydrogenase 1 (IDH1), with the latter resulting in a substitution of arginine for histidine at amino acid position 132 (R132H;).19 Functional studies have revealed that the H132 mutant protein catalyzes the conversion of α-ketoglutarate into 2-hydroxyglutarate (2-HG), a suspected oncometabolite.19,20 Mutant IDH1 was first reported in colorectal cancer21 and has been identified in acute myelogenous leukemia and in GBM through genome wide mutational analysis.22,23 A subsequent study found mutations in IDH1/2 in almost 80% of grades II and III astrocytomas, oligodendrogliomas and secondary GBMs,24 with the most common mutant being the G395A substitution. Microarray RNA profile analysis indicates that IDH1 mutations in gliomas occur primarily in the proneural subtype, making them distinctive from the classical, neural, and mesenchymal subtypes which rarely show this mutation25 and suggesting that this mutation is correlated to an improved prognosis and a potential stratifier for treatment options. Indeed, mutations in the IDH1 gene in all low glioma grades, as well as secondary GBMs, predict longer survival and better response to temozolomide treatment.26,27 Although several studies have reported that levels of 2-HG increase up to ~100-fold in glioma cell lines expressing mutant IDH1,28 a recent study failed to show a correlation between serum levels of 2-HG with IDH1/2 mutation status or tumor size in patients with glioma,29 suggesting that quantifying mutant mRNA copy number rather than the metabolite may provide more accurate information about tumor status.
Using BEAMing and ddPCR on mRNA from EVs, we reliably identified the mutant IDH1 mRNA in CSF-derived EVs collected at the time of brain tumor surgery from patients bearing mutant IDH1 glioma tumors, but not in serum-derived EVs. Under the conditions used, both assays gave similar levels of sensitivity and specificity in CSF samples. We also showed that CSF-derived EVs from patients with tumor have higher levels of IDH1 mRNA than CSF EVs from controls. A higher copy number of IDH1 mRNA was also found in serum from patients with tumor as compared with that from controls. In conclusion, we report that CSF-derived EVs provide a platform for detection of brain tumor specific biomarkers and that digital PCR platforms work well for detection of rare mutations in EV mRNAs.
Results
Isolation of serum and CSF-derived EVs for RNA-based mutation analysis
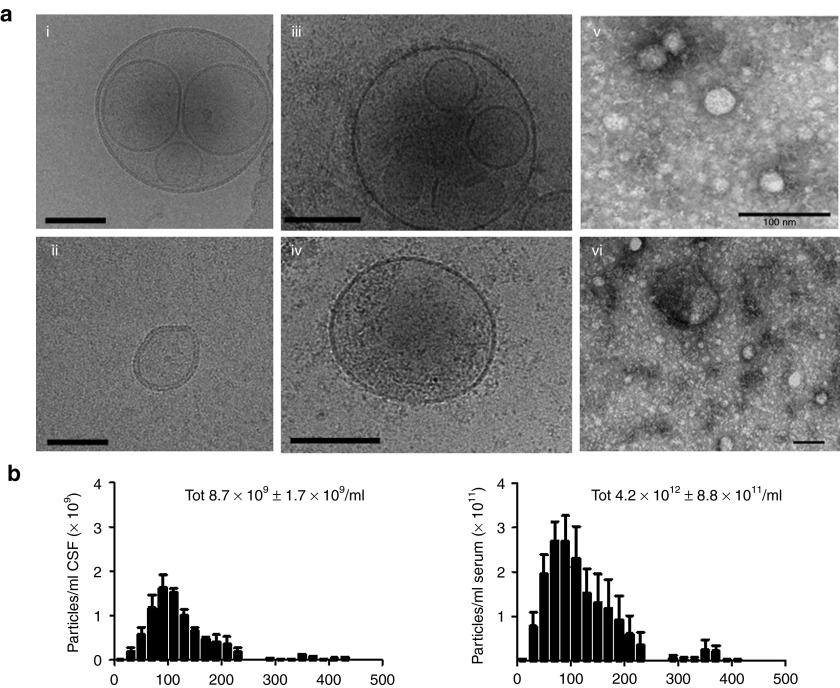
To prepare for EV-BEAMing (Figure 1) and ddPCR of EV RNA (Figure 2) analysis, we first isolated EVs from frozen biobanked serum and CSF of patients with grades II, III, and IV gliomas, as well as controls. Cryo-transmission electron microscopic and electron microscopic (EM) analysis of pelleted CSF-derived EVs and plasma, revealed round double-lipid membrane vesicles, sometimes with vesicles within vesicles and a size range of approximately 30–300 nm in diameter (Figure 3a). Nanosight tracking analysis revealed that GBM serum and CSF samples contained particles typically 30–300 nm in diameter, with an average of 4.2 × 1012 particles/ml ± 8.9 × 1011 SD (n = 3) in serum and 8.7 × 109 ± 1.7 × 109 SD particles/ml (n = 3) in CSF, making the particle number in CSF about 300-fold lower than in serum (Figure 3b). The quality of the extracted RNA from serum and CSF samples was assessed using the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA) and was confirmed to be similar to previously shown profiles3 (see Supplementary Figure S1). EV-derived RNA was then reverse transcribed into cDNA and 10% was used as input for RT-PCR. The remaining cDNA was preamplified (14 cycles) using dual-biotin–labeled forward primer and a reverse primer, used as input for EV-BEAMing as well as ddPCR and analyzed for IDH1 mRNA copy number and the G395 or A395 sequences in IDH1. FACS, ddPCR, and qPCR results were used to quantify the number of wild-type and mutant IDH1 mRNA copies present in EVs. In parallel, the tumor tissue was also typed for the A395 IDH1 mutation by genomic DNA sequencing.
Figure 1.
Overview of EV-BEAMing. EVs from serum or CSF samples were pelleted at 100,000g for 80 minutes and processed as follows: (1) RNA was extracted and (2) analyzed for total yield and quality using the Bioanalyzer (Agilent). (3) RNA was then reverse transcribed into cDNA and 1/2 of the sample was used to determine the IDH1 cDNA copy number inside EVs by (4) qPCR analysis. (5) The remaining sample was preamplified (14 cycles) and used as input for BEAMing PCR. The resulting DNA-coated beads were interrogated with sequence-specific fluorescent probes to produce beads with wild-type (green) and mutant (red) profiles. (6) The percentage of beads with mutant DNA was determined by FACS and used in conjunction with the qPCR data to determine the minimum number of copies present to allow reliable detection of the mutant message.
Figure 2.

Overview of droplet digital PCR. The RainDrop ddPCR workflow consisted of three steps: (1) PCR reaction mixtures for each sample were pipetted into one of eight wells on the Source microfluidic chip, which was next placed above empty PCR tubes in the RainDrop Source. Pressure-driven air flow forced the aqueous mixture into the chip along with a surfactant-containing fluorocarbon oil to generate five picoliter droplets, and then deposited automatically into the PCR tube strip. (2) The PCR tube strip was placed into a standard thermal cycler for endpoint PCR amplification, with single-target-molecule-containing droplets resulting in specific probe hydrolysis (PCR+) and bright fluorescence and the majority of droplets, containing no target molecule, resulting in only background probe fluorescence (PCR−). (3) Each droplet's fluorescence was detected using a Sense microfluidic chip. The fluorescence was detected and processed into a two-dimensional scatter plot display, custom software was used to draw appropriate gates for each droplet endpoint cluster, and the number of droplets within each gate was counted.
Figure 3.
Extracellular particle analysis using EM, cryoTEM and Nanosight. Cerebrospinal fluid (CSF), plasma and serum samples were collected upon or before the opening of the dura mater, respectively, processed as described in methods and stored at −80 °C until further processing. (a) CryoTEM of CSF EVs (i, ii), plasma EVs (iii, iv), and regular EM of CSF EVs (v, vi). Scale bars = 100 nm. (b) CSF (n = 3) and serum samples (n = 3) were diluted 1:50 and 1:3,000 respectively in PBS, and size and concentration of particles were assessed using nanoparticle tracking analysis (NanoSight; ±SD).
IDH1 mRNA copies in serum and CSF-derived EVs
EV mRNA was reverse transcribed into cDNA and used to perform copy number analysis for IDH1 transcripts using quantitative PCR. Serum from patients with glioma contained an average of 7,800 ± 6,600 SD (n = 10) copies of IDH1 mRNA/ml (Figure 4b), whereas serum from controls had 245 ± 65 SD (n = 4) copies of IDH1 mRNA/ml (Table 1; P ≤ 0.007). The copies of IDH1 mRNA in CSF from patients with glioma tended to be higher than in serum from patients with glioma, mean 106,000 ± 150,000 SD (n = 14) copies mRNA/ml in CSF (Figure 4b) and 7,800 ± 6,600 SD IDH1 mRNA/ml serum (P ≤ 0.03 n = 10). IDH1 mRNA copies were much lower in CSF from non-tumor controls (40 copies mRNA/ml; n = 2) as compared with CSF from patients with glioma (P ≤ 0.02). Note that all CSF glioma samples were collected from the CSF cisterns before tumor manipulation during surgery, except for glioma sample 308 and controls which were collected by lumbar puncture; this may partially explain the difference in IDH1 mRNA copy number. Interestingly, the lumbar puncture CSF sample 308 had the highest percentage of mutant IDH1 in the EVs.
Figure 4.
Correlation of samples characteristics with the detection of the mutant IDH1. (a) BEAMing FACS plot of one representative mutant positive samples indicating the mutant (Q1) and wild-type populations (Q4). (b) qPCR analysis of the IDH1 mRNA copy number in CSF (n = 14) and serum (n = 10) samples from all GBM patients analyzed that are wild-type or mutant for the IDH1 gene (±SEM). (c) Tumor volumes (left panel) and IDH1 mRNA copy numbers (right panel) in 1 ml of CSF from the mutant IDH1 glioma samples analyzed (n = 8), corresponding to detected or non detected, in mRNA as determined by EV-BEAMing assay and ddPCR.
Table 1. CSF and serum samples characteristics.

CSF EVs from patients with tumor carry the mutated IDH1 A395
For EV-BEAMing analysis, 50% of the IDH1 cDNA product was PCR amplified (14 cycles) and used as input for BEAMing as well as ddPCR, as described in the Materials and Methods. For BEAMing PCR, we interrogated the identity of the amplicons attached to each bead using an Alexa Fluor 488–labeled probe specific for the wild-type IDH1 sequence, an Alexa Fluor 647–labeled probe specific for the A395 mutant sequence, and a Pacific Blue (PB)–labeled control probe specific for a sequence common to both the mutant and wild-type sequences. This results in individual beads that are colabeled for PB (blue fluorescence) and either Alexa Fluor 488 (green fluorescence) or Alexa Fluor 657 (red fluorescence). FACS analyses were performed by initially gating on 100,000 (single bead) events which were PB-positive. Percentage mutant signal was calculated using the PB-positive population, thus, excluding beads that had no fluorescence and those with both wild-type and mutant fluorescence (Figure 4a; see Materials and Methods). Droplet digital PCR was also performed using the same probe sequences but for this assay they were labeled with FAM and VIC fluorescence (mutant and wild-type, respectively). After ddPCR amplification, the samples were transferred to a microfluidic chip for fluorescence measurements. Custom software was used to define gates and count the number of single molecule amplified droplets within each gate. For BEAMing PCR, the preamplified PCR products were diluted ~1:15,000 (10 pmol/reaction), whereas they were diluted 1:106 for ddPCR.
We analyzed the CSF of patients with grade II (n = 2), grade III (n = 4), and grade IV (n = 2) gliomas harboring the A395 mutation in IDH1, as well as that of grade II glioma (n = 1) and grade IV GBMs (n = 5) that possessed only wild-type IDH1 alleles and detected the mutant message in five out of eight of the mutant tumor samples and none of the wild-type samples (Figure 4c; Table 1). Quantitative PCR analysis revealed that the five mutant positive CSF samples had a wide range of copies of IDH1 message/cDNA from 1 ml of CSF (ID# 308, 2,262 copies; ID# 2244, 3,000 copies; ID# 2160, 5,210 copies; ID# 2199, 10,240 copies; and ID# 2510, 281,320 total copies of IDH1 message/cDNA, in the five samples respectively). The percentage of mutant message detected in the five samples was 8.0%, 5.0%, 1.5%, 0.4%, and 8.5%, respectively, suggesting that about 181, 150, 78, 41, and 23,912 amplifiable copies of the mutant message were obtained from EVs in 1 ml CSF from these patients. Neither of the two non-tumor control CSF samples, collected during routine neurological examination displayed any mutant events (Table 1). Droplet digital PCR also detected the same mutant samples but the percentage of the mutant message was generally twice as high; ID#s 308 = 23.7%, 2244 = 10.6%, 2160 = 3.9%, 2199 = 1.0%, and 2510 = 20.4%. The reasons for these platform differences are still being investigated. We also analyzed serum samples from seven patients with IDH1 mutant tumors (grades II, III, and IV), as well as four healthy control serum samples and the mutation was not detected in any of the seven IDH1 mutant serum samples or control samples (Table 1). Both digital PCR methods performed equally, although the dilutions of the preamplified PCR product differed greatly.
Discussion
Here, we show that the mutational profile of IDH1 mutated brain tumors can be measured in CSF without the need for a biopsy of the tumor. We can measure the presence as well as the abundance of the mutated transcript found in CSF EVs with digital PCR methods such as BEAMing or ddPCR. This method may be useful as a non-invasive tool to verify and subtype brain tumors in cases where the tumor location makes biopsies risky or impossible. It may also be used to subtype tumors for drug clinical trial enrollment. BEAMing and ddPCR assays had the same sensitivity and specificity levels using CSF. Serum contains vesicles from many normal processes naturally occurring in the blood such as maturation of reticulocytes and platelet activation. The CSF may therefore be a more enriched source of the brain tumor–derived vesicles and therefore easier to detect than in serum. Interestingly, the number of copies of IDH1 mutant mRNA in CSF EVs from patients with tumor was significantly higher than in controls (Table 1). Noteworthy is the difference in sampling site, most CSF samples from patients with tumor were collected upon opening of the dura mater, near the primary tumor, whereas CSF control samples were collected by lumbar puncture. Patient sample 308 was collected by lumbar puncture and shows that mutation detection can be done from lumbar puncture, and this sample actually had the highest percentage of mutant IDH1 transcripts. The high levels of IDH1 mRNA in cisternal CSF from patients with glioma collected during surgery compared with control patients collected by lumbar puncture may be due to a higher number of EVs released by tumor or normal cells in the vicinity of the tumor or from debris from normal cells in the mechanical process of opening the dura mater. Two of the false negative samples (in the EVs) had unusually high copy numbers of WT IDH1 (~400,000 WT copies; Table 1) that could have masked the detection of the mutant. For this purpose, lumbar puncture samples may be preferred for this analysis.
Using the BEAMing assay we determined that between 41 and 181 mutant IDH1 mRNA transcripts were present in 1 ml CSF in four samples, with one sample having as many as 23,912 mutant copies. ddPCR detected roughly twice as many mutated RNA transcripts per ml CSF (ID#s 308 = 536, 2244 = 318, 2160 = 203, 2199 = 102, and 2510 = 57,389). Interestingly, two of the three IDH1 mutant positive tumors that were not detected by either method were grade II CSF samples (IDs# 2287 and 2516, Table 1). The third sample that was not detected was a grade III tumor (ID# 2513) that had the smallest tumor size in grade III category suggesting a possible correlation with grade as well as tumor size (Figure 4c). Further studies with a larger number of samples will address this correlation more thoroughly. The IDH1 mutant detection sensitivity in serum will need further optimization, and the relatively low IDH1 mRNA copy number suggest that increasing the volume of serum input may increase the sensitivity of the assay. All tumor-free controls included in the study were wild-type for IDH1 mRNA as analyzed by FACS (Table 1). Future studies will aim to increase the copy number input to improve the sensitivity of the assay in blood-based assays and to correlate the detection of mutant IDH1 with clinical, demographic, MRI and molecular neuropathologic studies. Other studies have reported detection of other tumor biomarkers in blood on DNA, such as the EGFR gene amplification in lung adenocarcinomas30 and PIK3CA mutations in metastatic breast cancer.31 We have previously shown that also DNA can be present in glioma EVs;3 however, this study focused only on the RNA fraction.
IDH1 is usually a heterozygous mutation,28 so when 5% of the IDH1 transcripts are mutated, approximately 10% of the IDH1 transcripts come from the tumor. It may be possible to enrich the tumor EV fraction by immunoaffinity methods using antigens specific to the tumor cell surface32 or using magnetic resonance detection of mutant proteins to potentially increase the sensitivity of mutation assays.33
Information on the mutational profiles of different types of gliomas has increased to the point where genomic alterations and expression profiles characteristic of the classical, proneural, neural, and mesenchymal glioma subtypes have been identified.12 EV-BEAMing or ddPCR on biofluid EVs should have high value in monitoring specific subtypes of gliomas in a minimally invasive manner. Although obtaining CSF is not a routine procedure in patients with brain tumors, it is markedly less invasive than obtaining an intracerebral biopsy of the tumor. Further extension of this work in clinical studies will require focusing on samples derived by lumbar puncture, as this is the only feasible sampling methodology to perform on a routine basis but this study lays the foundation for lumbar puncture–based CSF mutation analysis studies in patients. This technology should enable monitoring of a variety of mutations and transcript levels in tumors using CSF and potentially serum-derived EVs not only when the tumor is located in areas with inherent risk of sampling such as brain, but also for tumors in other parts of the body. This study provides a proof of principle for detection of single base pair mutations in tumor RNA using EVs in CSF samples, combined with a sensitive digital PCR assays.
Materials and methods
Study design and sample collection. This study was approved by the Institutional Review Boards of Massachusetts General Hospital, Beth Israel Deaconess Medical Center, and the University of California, Los Angeles. Serum and CSF samples from patients and healthy volunteers were collected according to approved IRB protocols. Brain tumors were obtained at the Massachusetts General Hospital, Beth Israel Deaconess Medical Center and University of California, Los Angeles and confirmed by histopathology. The mutational status of the IDH1 gene in tumor tissue was determined by genomic sequencing or SNaPShot analysis by the Molecular Neuropathology Laboratory at UCLA and Massachusetts General Hospital, respectively.
Sample processing. Blood was collected from patients undergoing surgical removal of gliomas using BD Vacutainer SST tube (BD Biosciences, San Jose, CA), before opening of the dura mater, maintained for 30 minutes at room temperature and then centrifuged at 1,300g for 10 minutes. The serum was separated from the coagulated blood cells, filtered through a 0.8 μm filter and stored at −80 °C. CSF, also collected at surgery from patients with glioma upon opening of the dura mater, was centrifuged at 300g for 10 minutes to remove cells, and stored at −80 °C. Control CSF samples were collected by lumbar puncture from patients with suspected non-malignant, neurological disease whose CSF profile was later found to be normal, and stored at −80 °C. At the time of analysis, samples were thawed, centrifuged at 100,000g for 80 minutes at 4 °C to collect EVs and RNA was extracted, as previously described.3 To minimize any risk of contamination, tumor tissue was extracted and sequenced in one laboratory, whereas serum and CSF EV RNA was extracted at a different location in a DNA-free, RNA extraction facility, and BEAMing and ddPCR were performed at a third location.
Extravesicular RNA isolation. Frozen serum or CSF samples were thawed (1 ml/sample), diluted to 2.5 ml with sterile PBS and centrifuged at 100,000g for 80 minutes. External DNA on the EVs was removed using 2 µl of DNAse from the Turbo DNA-free kit (Ambion, Foster City, CA), according to manufacturer's recommendations. RNA was purified using the miRNeasy Qiagen kit (Qiagen, Valencia, CA) and eluted in 30 µl RNase-free water. RNA quality was assessed using a 2100 Bioanalyzer (Agilent) with an RNA 6000 Pico Chip kit (Agilent).
Determination of tumor volume. For high grade gliomas with significant contrast enhancement, T1-weighted sequences after injection of contrast agent were used to define the “tumor volume” as the area of contrast enhancement including central necrotic tissue. For low grade tumors and tumors without significant enhancement, volume was determined using T2 areas of hyperintensity. To determine the volume, we measured the tumor area in each slice using a region-of-interest method (BrainLAB Software; BrainLAB, Feldkirchen, Germany). The volumes were calculated by multiplying these resulting areas by the slice thickness. The total tumor volume is the sum of the volumes calculated for each of the multiple parallel slices.34
Nanosight tracking analysis. Nanosight tracking analysis was used to count and size EVs. Serum was processed as above and stored at −80 °C. At the time of the analysis the serum was diluted 1:5,000 and analyzed using the Nanosight. CSF was also stored at −80 °C and diluted 1:50 for Nanosight analysis. Both biofluids were diluted in PBS for nanosight tracking analysis.
Electron microscopy. CSF samples (500 μl) were thawed and ultracentrifuged at 100,000g for 80 minutes. The EV pellet was then resuspended in 50 μl of PBS and analyzed by EM. 2.5 μl of the samples was adsorbed for 30 minutes on carbon coated grids that had been made hydrophilic by a 30-second exposure to a glow discharge. Excess liquid was removed with a Whatman filterpaper (GE Healthcare, Piscataway, NJ) and the samples were stained with 0.75% uranyl formate for 30 seconds. After removing the excess uranyl formate with a filter paper the grids were examined in a JEOL 1200EX Transmission EM or a TecnaiG2 Spirit BioTWIN and images were recorded with an AMT 2k CCD camera.
Cryo-transmission EM. CSF and plasma samples were preserved in vitrified ice supported by holey carbon films on 400-mesh copper grids. Each sample was prepared by applying a 3 µl drop of sample suspension to a cleaned grid, blotting away with filter paper, and immediately proceeding with vitrification in liquid ethane. Grids were stored under liquid nitrogen until transferred to the EM for imaging. EM was performed using an FEI Tecnai T12 EM, operating at 120keV equipped with an FEI Eagle 4k x4k CCD camera. Vitreous ice grids were transferred into the EM using a cryostage that maintains the grids at a temperature below −170 °C. Images on each grid were acquired at multiple scales to assess the overall distribution of the specimen. After identifying potentially suitable target of areas for imaging at lower magnifications, pairs of high magnification images were acquired at normal magnifications of 110,000×, 52,000×, and 21,000×.
Quantitative RT-PCR. mRNA was reverse transcribed into cDNA using the SuperScript VILO cDNA synthesis kit (Invitrogen, Carlsbad, CA). To determine the copy number of IDH1 transcripts, we performed quantitative PCR on the cDNA. Reactions were done in a 25 µl reaction using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and 160 nmol/l of each primer. Amplification conditions consisted of: (i) 1 cycle of 50 °C, 2 minutes; (ii) 1 cycle of 95 °C, 10 minutes; (iii) 40 cycles of 95 °C, 15 seconds; and 60 °C, 1 minute, and (iv) a dissociation stage consisting of 1 cycle of 95 °C, 15 seconds; 60 °C, 20 seconds; and 95 °C, 15 seconds on the 7000 ABI Prism PCR system (Applied Biosystems). Ct values were analyzed in auto mode and manually inspected for accuracy. Primer dimers were excluded by evaluation of dissociation curve and agarose gel electrophoresis. The following IDH1 primers were used: Forward: CGGTCTTCAGAGAAGCCATT and Reverse: AGGCCCAGGAACAACAAAAT.
Pre-amplification of IDH1 cDNA. We performed IDH1 PCR on the cDNA derived from EV RNA using forward (5′-tcccgcgaaattaatacgacCGGTCTTCAGAGAAGCCATT-3′) and reverse (5′-gctggagctctgcagctaAGGCCCAGGAACAACAAAAT-3′) primers flanking nucleotide position 395 of the IDH1 transcript, where lower case letters denote universal primer sequences and upper case letters denote IDH1 specific primer sequences. Ten μl of cDNA was split into two PCR reactions, each performed in 50 μl volumes containing 5 μl cDNA, 1 U of Phusion Hot Start II DNA Polymerase (New England Biolabs, Ipswich, MA), 1X Phusion HF buffer (New England Biolabs), 0.2 mmol/l dNTP mix (Invitrogen), and 0.2 μmol/l of each primer. Amplification was done in a Veriti Thermal Cycler (Applied Biosystems) using the following conditions: 98 °C for 3 minutes; 10 cycles of 98 °C for 10 seconds, 68 °C for 15 seconds (−0.5 °C/cycle), 72 °C for 10 seconds; 30 cycles of 98 °C for 10 seconds, 62 °C for 15 seconds, 72 °C for 10 seconds; 1 cycle of 72 °C for 5 minutes. The PCR products for each sample were then pooled and purified using a QIAquick PCR purification column (Qiagen) and the concentration determined using a 2100 Bioanalyzer with a DNA 1000 LabChip kit (Agilent).
EV-BEAMing PCR. PCR amplicons from each sample were used as templates for each BEAMing analysis. We performed the BEAMing PCR using a protocol slightly modified from that previously described.11 We set up 150 μl PCR reactions containing 10 μl 20 pmol/l template DNA, 2X TITANIUM Taq DNA Polymerase (Clontech, Mountain View, CA), 1X TITANIUM Taq buffer (Clontech), 1 mmol/l dNTP mix, 8.3 mmol/l MgCl2 (Invitrogen), 0.05 μmol/l universal forward primer (5′-tcccgcgaaattaatacgac-3′), 8 μmol/l nested reverse primer (5′-AATCAGTTGCTCTGTATTGATCC-3′), and 6 × 107 magnetic streptavidin beads (MyOne, Invitrogen) coated with a modified universal forward primer (5′-dual biotion-T-Spacer18-tcccgcgaaattaatacgac-3′). The aqueous reaction mixture was then combined with 600 μl of an oil/emulsifier mixture of 7% w/v ABIL WE09 (Degussa, Essen, GE), 20% v/v M3516 mineral oil (Sigma-Aldrich, St Louis, MO) and 73% v/v Tegosoft DEC (Degussa), and one 5 mm steel bead (Qiagen) in one well of a 96-deep-well plate (Abgene, Lafayette, CO). The plate was shaken in a TissueLyser (Qiagen) for 10 seconds at 15 Hz and then 7 seconds at 17 Hz, and 80 μl of the emulsions were distributed into each well of a MicroAmp 96-well PCR plate (PE Biosystems, Carslbad, CA). The emulsion PCR was done in a Veriti Thermal Cycler set to a 20% temperature ramp rate to prevent disruption of the emulsions, with the following cycling conditions: 94 °C for 3 minutes; 12 cycles of 94 °C for 10 seconds, 65 °C for 45 seconds (−0.5 °C/cycle), 70 °C for 75 seconds; 50 cycles of 94 °C for 10 seconds, 59 °C for 45 seconds, 70 °C for 75 seconds; 1 cycle of 70 °C for 5 minutes. We broke the emulsions by adding 150 μl breaking buffer (10 mmol/l Tris–HCl, pH 7.5, 1% Triton X-100, 1% SDS, 100 mmol/l NaCl, 1 mmol/l EDTA) to each well and mixing the samples with a TissueLyser (Qiagen) at 20 Hz for 30 seconds. The samples were then centrifuged at 3,200g for 2 minutes and the oil phase was removed. We performed this breaking procedure twice and then combined and resuspended all of the beads for each respective sample in 100 μl wash buffer (20 mmol/l Tris–HCl, pH 8.4, 50 mmol/l KCl). The beads were incubated with 0.1 mol/l NaOH to remove non-biotinylated DNA, and then washed and resuspended in 100 μl wash buffer.
Mutational analysis. We interrogated the mutational status of the bead-bound DNA using three fluorescent oligonucleotides: an Alexa Fluor 488 (AF488)–labeled oligonucleotide (5′-AF488-ATAAGCATGACGACCTATGAT-3′) specific to the wild-type IDH1 sequence, an Alexa Fluor 647 (AF647)–labeled oligonucleotide (5′-AF647-ATAAGCATGATGACCTATGAT-3′) specific to the A395 IDH1 variant, and a PB-labeled control oligonucleotide (5′-PB-TTTTACCCATCCACTCACAA-3′) specific to a sequence region shared by both wild-type and mutant IDH1 variants. The oligohybridization was in a 100 μl hybridization mixture of 30 μl beads, 66 μl hybridization buffer (3 mol/l tetramethylammonium chloride (Sigma-Aldrich), 50 mmol/l Tris–HCl, pH 7.5, 4 mmol/l EDTA) and 4 μl of a mixture of the oligonucleotides, each at 5 μmol/l. Samples were heated to 70 °C for 10 seconds, cooled to 35 °C at 0.1 °C/second, incubated at 35 °C for 2 minutes, and finally cooled to 25 °C at 0.1 °C/second using a Veriti Thermal Cycler. The beads were collected, washed in 100 μl hybridization buffer at 48 °C for 5 minutes, and then 100 μl wash buffer at room temperature, before a final resuspension in 400 μl wash buffer. We analyzed all oligohybridized beads using an LSR II flow cytometer (BD Biosciences) and processed the data using FACSDiva software (BD Biosciences). Events were gated to exclude doublets, aggregates, and beads without IDH1 amplicons. At least 100,000 events were counted and the mutant signal was calculated using events that had either wild-type or mutant fluorescence, but not both.
Digital droplet PCR methods.
Reaction preparation: Pre-amplified PCR product was diluted and added to the reaction mixture. Primers and fluorescent Taqman MGB probes (FAM-labeled G395A-5′- ATAAGCATGATGACCTATGAT-3′ and VIC-labeled WT- ATAAGCATGACGACCTATGAT-3′) were designed using Applied Biosystems' Custom TaqMan Assay Design Tool. The final IDH1 assay mix contained 900 µmol/l forward and reverse primers, 200 nmol/l VIC-labeled WT IDH1 probe, and 200 nmol/l FAM-labeled G395A probe. The PCR reaction was prepared in 25 µl final volume, using 12.5 µl TaqMan Genotyping Master Mix (Life Technologies), 2.5 µl Droplet Stabilizer (RainDance Technologies, Lexington, MA), 2.5 µl Assay Mix (10×) and 7.5 µl cDNA template in water.
Digital droplet assay: Droplets containing PCR reaction components were generated using a hydrodynamic flow-focusing microfluidic chip (RainDrop Source chip; RainDance Technologies), deposited into PCR tubes as 5 pl aqueous droplets suspended in inert fluorinated oil (REB Carrier Oil; RainDance Technologies), and subjected to PCR amplification in a thermal cycler (Mastercycler pros; Eppendorf, Hamburg, Germany). PCR cycling parameters: 10 minutes 95 °C, then 44 cycles of 95 °C for 15 seconds and 60 °C for 1 minute. After PCR completion, the emulsion was injected into a second microfluidic chip (RainDrop Sense chip; RainDance Technologies) for fluorescence measurement. Following 488 nm excitation, droplet fluorescence was detected through filters (FAM and VIC centered emission) with photomultiplier tubes recording fluorescence intensity (“height”) and duration (“width”). Spectral crosstalk-corrected data from each sample or control were converted to a two-dimensional (FAM and VIC intensity) histogram (electronic and photonic noise was removed, droplet data were width filtered and normalized to background fluorescence). Custom software (RainDrop Analyst; RainDance Technologies) was used to define graphical areas or “gates” associated with each allele type and count the number of droplets within each gate.
SUPPLEMENTARY MATERIAL Figure S1. RNA profiles from CSF and serum EVs.
Acknowledgments
This work was supported by NIH/NCI grants CA069246 (F.H., X.O.B., B.C.); CA141226 (X.O.B.); CA156009 (X.O.B.); and CA141150 (X.O.B.); Brain Tumor Funders' Collaborative (B.C., X.O.B.); American Brain Tumor Association (ABTA; J.S.); Harvard Catalyst (B.C.); A Reason To Ride research fund (E.T.W.); Hyugens Scholarship NL (L.B.), the Richard Floor Biorepository and the Accelerate Brain Cancer Cure (ABC2). All processing of samples by Exosome Diagnostics Inc and RainDance Technologies was carried out on de-identified samples. We thank Suzanne McDavitt for skilled editorial assistance. We thank N Agrawal for his advice and for providing us with BEAMing reagents and A Mandinova for providing access to the TissueLyser. We thank Karen Messer for help with statistical analysis. We also thank Winston Patrick Kuo and Fatemeh Momen-Heravi for help with TEM and Mikkel Noerholm for help with probe design. J.S. is an inventor on the microvesicle technology used in this study which has been licensed to Exosome Diagnostics, Inc. He holds equity in, and is an employee of that company. Breakefield and Carter are on the Scientific Advisory Board of the company for which they receive cash compensation. M.L.S. and S.K.K. are employees of RainDance Technologies Inc. The other authors declared no conflict of interest.
Supplementary Material
RNA profiles from CSF and serum EVs.
References
- Lecomte T, Ceze N, Dorval E, Laurent-Puig P. Circulating free tumor DNA and colorectal cancer. Gastroenterol Clin Biol. 2010;34:662–681. doi: 10.1016/j.gcb.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noerholm M, Balaj L, Limperg T, Salehi A, Zhu LD, Hochberg FH, et al. RNA expression patterns in serum microvesicles from patients with glioblastoma multiforme and controls. BMC Cancer. 2012;12:22. doi: 10.1186/1471-2407-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong BS, Cho JH, Kim H, Choi EJ, Rho S, Kim J, et al. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics. 2009;10:556. doi: 10.1186/1471-2164-10-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Vizio D, Kim J, Hager MH, Morello M, Yang W, Lafargue CJ, et al. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009;69:5601–5609. doi: 10.1158/0008-5472.CAN-08-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS, et al. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med. 2012;10:5. doi: 10.1186/1479-5876-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston CL, Glantz MJ, Connor JR. Detection of cancer cells in the cerebrospinal fluid: current methods and future directions. Fluids Barriers CNS. 2011;8:14. doi: 10.1186/2045-8118-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelwell N, Millington S, Solinas A, Booth J, Brown T. Mode of action and application of Scorpion primers to mutation detection. Nucleic Acids Res. 2000;28:3752–3761. doi: 10.1093/nar/28.19.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3:551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chen WD, Papadopoulos N, Goodman SN, Bjerregaard NC, Laurberg S, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27:858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Schmidt K, Durkee KH, Moore KJ, Goodman SN, Shuber AP, et al. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology. 2008;135:489–498. doi: 10.1053/j.gastro.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taly V, Pekin D, El Abed A, Laurent-Puig P. Detecting biomarkers with microdroplet technology. Trends Mol Med. 2012;18:405–416. doi: 10.1016/j.molmed.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Pekin D, Skhiri Y, Baret JC, Le Corre D, Mazutis L, Salem CB, et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip. 2011;11:2156–2166. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Bhattacharya S, Kotsopoulos S, Olson J, Taly V, Griffiths AD, et al. Multiplex digital PCR: breaking the one target per color barrier of quantitative PCR. Lab Chip. 2011;11:2167–2174. doi: 10.1039/c1lc20126c. [DOI] [PubMed] [Google Scholar]
- Didelot A, Kotsopoulos SK, Lupo A, Pekin D, Li X, Atochin I, et al. Multiplex picoliter-droplet digital PCR for quantitative assessment of DNA integrity in clinical samples. Clin Chem. 2013;59:815–823. doi: 10.1373/clinchem.2012.193409. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Cancer Genome Atlas Research Network et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labussière M, Idbaih A, Wang XW, Marie Y, Boisselier B, Falet C, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74:1886–1890. doi: 10.1212/WNL.0b013e3181e1cf3a. [DOI] [PubMed] [Google Scholar]
- SongTao Q, Lei Y, Si G, YanQing D, HuiXia H, XueLin Z, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012;103:269–273. doi: 10.1111/j.1349-7006.2011.02134.x. [DOI] [PubMed] [Google Scholar]
- Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29:6409–6417. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- Capper D, Simon M, Langhans CD, Okun JG, Tonn JC, Weller M, German Glioma Network et al. 2-Hydroxyglutarate concentration in serum from patients with gliomas does not correlate with IDH1/2 mutation status or tumor size. Int J Cancer. 2012;131:766–768. doi: 10.1002/ijc.26425. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res. 2011;17:7808–7815. doi: 10.1158/1078-0432.CCR-11-1712. [DOI] [PubMed] [Google Scholar]
- Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;18:3462–3469. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010;10:505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi WM, Wildrick DM, Sawaya R. Volumetric measurement of brain tumors from MR imaging. J Neurooncol. 1998;37:87–93. doi: 10.1023/a:1005944724470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA profiles from CSF and serum EVs.